Abstract
The plausibility of sympatric speciation is still debated despite increasing evidence, such as host races in insects. This speciation process may be occurring in the case of the two phenological forms of the obligatorily myrmecophilous Phengaris arion. The main goal of our research was to study the nature and causes of difference between these forms focusing primarily on the incipient speciation via host races. Molecular analyses based on highly variable microsatellites together with Wolbachia screening, male genitalia morphometrics and host ant studies were carried out on four syntopic sample pairs. Our results show that the two phenological forms of P. arion may meet the criteria for host plant races. They coexist in sympatry in certain parts of the species range which is allowed by the adaptation to the distinct phenology of the host plants. Negative selection acts against the intermediate individuals which are on the wing in the inappropriate time frame. Thus, disruptive selection affects and produces bimodal distributions of phenotypes. However, the phenology of food plants is not entirely distinct and fluctuates year by year. Therefore, the two forms can exchange genes occasionally depending on the length of the time slot when they can meet with each other. Consequently, the reproductive isolation could not be completed and the existence of the two arion forms may represent only an incipient stage of sympatric speciation. It is also clear that Wolbachia is likely not a driver of sympatric speciation in this case.
Similar content being viewed by others
Introduction
Speciation is considered as a more or less continuous process starting from polymorphisms within species through the emergence of intermediate forms with increasing reproductive isolation up to the formation of new species. However, the exact route to speciation remains contested in many cases (Turelli et al. 2001). Traditionally, allopatric speciation is regarded as a null model of speciation, which requires only geographical isolation and time long enough (Futuyma and Mayer 1980). While the opposite extreme, sympatric speciation is divergence within a single geographical region such that the range of one nascent species completely overlaps the other (Fitzpatrick et al. 2008). Gavrilets (2003) pointed out that such a geographical definition is not precise enough because it does not specify the population structure of the ancestral population. Therefore, evolutionary biologists have redefined sympatric speciation non-spatially to require panmixia between a pair of demes before onset of reproductive isolation (Mallet et al. 2009). Although sympatric speciation is possible even in this restricted population genetic sense (Fitzpatrick et al. 2009), it is still thought by many to be rare in its most general sense.
Substantial indications, however, now exist that sympatric speciation may have a significant role in the evolution of insects (Berlocher and Feder 2002; Bolnick and Fitzpatrick 2007; Coyne 2007; Drès and Mallet 2002; Via 2001). Host-associated biotypes, including host races in plant-feeding insects, have often been used as evidence for the plausibility of sympatric speciation representing its incipient stage (Bush 1969; Feder 1998; Filchak et al. 2000; Tauber and Tauber 1989). Host races are defined as sets of populations that (i) coexist in sympatry in at least part of their range, (ii) use different host taxa and consist of individuals that exhibit ‘host fidelity’, i.e. are associated with a particular host, (iii) are genetically differentiated at more than one locus, (iv) regularly exchange genes at a rate of more than ca. 1% per generation and (v) are spatially and temporally replicable, i.e. are more genetically differentiated from populations on another host in sympatry (and at the same time) than at least some geographically distant populations on the same host (Drès and Mallet 2002).
Sympatric speciation through these intermediates involves ecologically driven reproductive isolation associated with adaptation to alternative discrete resources and/or niches. Selection against intermediate phenotypes is the central driving force for sympatric speciation (Nosil 2012). These intermediates are disadvantageous because they accrue fewer resources as a result of density- and frequency-dependent selection or they procure fewer mates as a result of preferences for extreme phenotypes (Turner and Burrows 1995). Disruptive selection may produce bimodal distributions of phenotypes or drive the evolution of reproductive isolation for sympatric taxa which are then maintained usually via assortative mating (Dieckmann and Doebeli 1999; Turelli et al. 2001).
The existence of different host races together with the possibility of sympatric speciation has emerged in the case of the obligatorily myrmecophilous Large Blue Phengaris arion (Linnaeus, 1758). This species has a very special socially parasitic life cycle depending on the dual presence of the specific initial food plant and host ant species. Females lay their eggs on flower-buds of their specific initial food plant. Young larvae feed on developing seeds, quickly growing through three instars but gaining only a few percent of their final weight. After 2–3 weeks, larvae drop to the ground and wait for foraging Myrmica ant workers which adopt them. In the ant nest, larvae follow a ‘predatory’ strategy, preying on ant brood for 10–11 months (Thomas et al. 1989). The average life span of imagoes is only a few days (Nowicki et al. 2005; Osváth-Ferencz et al. 2017).
Previous genetic studies have investigated a wide variety of P. arion populations concerning the biotope, larval food plant and host ants of the butterfly (Sielezniew et al. 2015; Sielezniew and Rutkowski 2012). According to these studies, the pattern of ecological variation does not influence the current genetic structure. Instead of this, it is probably shaped by landforms and the recent isolation of populations related to habitat fragmentation but local ant-related ecological specializations may be also a potential factor. However, these surveys did not aim to investigate the temporal dynamics of differentiation related to phenological variation of the target species.
P. arion has two phenological forms whose coexistence has been recorded in certain parts of the species range, such as the Carpathian Basin (Bereczki et al. 2011, 2014, 2015). The fast-flying, smaller-sized and dark violet-blue form (referred to as ‘spring arion’ hereafter) flies from mid-May to mid-June and prefers short-grass dry swards with cushions of early flowering Thymus species, which serve as initial food plants. The slower, larger and light silvery blue form (referred to as ‘summer arion’) is on the wing from the end of June to mid-August and mostly occurs at xerothermic oak forest fringes, on woodland clearings and in fen-like habitats in hilly areas. Females oviposit among flower-buds of late-flowering Thymus species and/or Origanum vulgare L.
Although previous studies have found significant morphological differences between the two arion forms both in wing and genitalia, they could not show any genetic isolation between them based on either the investigated mitochondrial and nuclear gene regions or allozyme loci (Bereczki et al. 2011, 2014, 2015). Nevertheless, the authors raised the possibility that the analysed markers were not suitable for the detection of the divergence between the forms because of their fairly low variability. They hypothesised that all extant differences between the spring and the summer arion are attributable to (1) different host-ant use, (2) incipient speciation, (3) cytoplasmic incompatibility induced by Wolbachia, or a combination of these factors.
Intracellular bacteria belonging to the genus Wolbachia have attracted considerable interest in the past decade primarily because of their vast abundance and fascinating effects on their hosts, which range from reproductive manipulation to mutualism (Werren et al. 2008). They are usually vertically transmitted from mother to offspring manipulating the host reproduction in ways that enhance their transmission to future generations. Accordingly, Wolbachia infections are associated with a variety of phenotypic effects on the hosts, such as cytoplasmatic incompatibility (CI) when the sperm of infected males is incapable of fertilizing the eggs of females which are uninfected or infected with a different strain (Hoffmann and Turelli 1997; Werren et al. 2008). The induction of CI between diverging populations could drive the evolution of new species. Therefore, Wolbachia may have an important role in accelerating the evolution of their hosts (Werren 1997). However, numerous studies also exhibit a positive correlation between bacterial titer and CI highlighting the importance of Wolbachia density for CI expression (Bourtzis et al. 1996; Boyle et al. 1993; Breeuwer and Werren 1993; Calvitti et al. 2015; Ikeda et al. 2003; Veneti et al. 2003). Beyond maternal transmission Wolbachia can also spread horizontally between phylogenetically distant but ecologically linked species (Hurst et al. 1992; Van Meer et al. 1999; Werren et al. 1995; Zhou et al. 1998). There are well-known examples when predacious species bear the same Wolbachia strain as their prey (Hoy and Jeyaprakash 2005; Kittayapong et al. 2003; Le Clec’h et al. 2013). This transmission route could have importance in P. arion as its caterpillars feed on the larvae of the host ants. Multiple host ant use provides the butterfly an opportunity to obtain a variety of bacterial strains which may induce CI between the competing strains driving ultimately to speciation in their butterfly host.
Here, we study the nature and causes of difference between the two phenological forms of P. arion focusing primarily on incipient speciation via host races. Therefore, we aimed (i) to estimate the level of genetic differentiation and gene-flow between the two forms of P. arion on the basis of highly variable microsatellites, (ii) to reveal whether differences in external genital traits also emerge between sympatric populations of the different forms systematically, (iii) to gather information on possible differences in host ant use of spring and summer arion and (iv) to estimate the intracellular Wolbachia infestation both in butterflies and their ant hosts.
Materials and methods
Samples
We sampled four syntopic pairs of the two P. arion forms in the Aggtelek Karst region of NE Hungary (Fig. 1), namely on the Korlát hill (TKo and NKo), Zabanyik (TZa and NZa), Perkupa (TPk and NPk) and Szin (TSi and NSi). The letter ‘T’ in the sample codes refers to spring arion, the letter ‘N’ indicates summer arion. Altogether, 138 adults were collected out of which 68 were spring and 70 were summer arion (see Suppl. Table S1 for more details). The two arion forms were differentiated based on the date of collection and their external characters. Imagoes were collected with a butterfly net and stored as dried material until molecular analyses.
Microsatellite studies
DNA was extracted from the proximal part of the abdomen following the protocol in Bereczki et al. (2014). Microsatellite polymorphism was studied at 9 loci, namely Macu8, Macu11, Macu15, Macu44, Macu45, Macari05, Macari16, Macari19 and Macari22 characterised by Ugelvig et al. (2011, 2012) and Zeisset et al. (2005). During amplification, we used fluorescent dye-labelled primers described by the authors mentioned above, and PCR reagents and conditions described in Rácz et al. (2015). After amplification, microsatellite products were multiplexed in two reactions (multiplex 1 with the loci Macu11, 15, 44, 45, Macari16 and multiplex 2 with the other four loci) and fragment analysis was carried out on an ABI 3130 Genetic Analyser in the Molecular Taxonomy Laboratory of the Hungarian Natural History Museum (Budapest, Hungary). Allele sizes were estimated using Peak Scanner software (Thermo Fisher Scientific, Waltham, MA, USA). Micro-Checker 2.2.3 (Van Oosterhout et al. 2004) was used for calculating null allele frequency by Monte Carlo simulation of expected homozygote frequencies and heterozygote allele size differences.
Parameters of polymorphism, including the average number of alleles per locus (Na), the effective numbers of alleles (Ne), allelic richness (AR), Shannon’s information index (I), proportion of polymorphic loci on the basis of the 95% criterion (P%), observed heterozygosity (Ho), expected heterozygosity (He) and Wright-index for heterozygote deficiency (FIS) values were calculated for both forms using GenAlEx 6.502 (Smouse et al. 2015) and Fstat 2.9.3.2 (Goudet 1995).
Genetic differentiation between the two types of P. arion was analysed using FST values (Weir 1996; Wright 1978). On the one hand, pairwise FST values were calculated among populations using the ‘hierfstat’ package (Goudet 2005) in the R computing environment (R Core Team 2018). Based on the obtained FST matrix, we constructed an unrooted neighbour joining (NJ) tree using the ‘ape’ package (Paradis et al. 2004). On the other hand, we computed FST values per locus between the syntopic sample pairs by Fstat 2.9.3.2 (Goudet 1995).
Analysis of molecular variance (AMOVA) was performed using Arlequin 3.5.1.2 (Excoffier and Lischer 2010). In the computation of hierarchical AMOVA, the samples were grouped according to their phenology (spring vs. summer type) or their geographical origin (i.e. locality). In this analysis, the total genetic variation was partitioned into four components: between groups, between phenology within locality or between locality within phenology (depending on the grouping), between samples within a group and within samples.
The genetic structure of the populations was analysed using Bayesian-clustering method (Pritchard et al. 2000). Here, we estimated the most probable number of genetically differentiated groups (K) in our populations and assigned the individuals to these groups. Structure 2.3.4 was run to carry out these analyses using default settings with an initial burn in of 100,000 steps and running length of 500,000 steps. In the evaluation of the results, ΔK was computed which indicates the change in log probability between successive K values (Evanno et al. 2005). Structure Harvester Web 0.6.93 (Earl and Von Holdt 2012) was used to compute the ΔK values.
The number of effective migrants (Nm) was also estimated between samples by Barton and Slatkin’s method (Barton and Slatkin 1986), which based on private alleles, using Genepop on the web (Raymond 1995; Rousset 2008).
Wolbachia studies
The same DNA extracts used for microsatellite studies were screened for Wolbachia. Each specimen was screened by the amplification of the highly conservative 16S ribosomal RNA gene with Wolbachia specific W-Spec primers developed by Werren and Windsor (2000). The amplification procedure described in Rácz et al. (2015) was followed. We used positive (confirmed infected samples) and negative controls (master mix without any DNA sample) in each reaction. The success of PCRs, i.e. Wolbachia presence, was checked by running 2 μl of product on 1% agarose gels stained with GelRed Nucleic Acid Stain (Biotium Inc., Fremont, CA, U.S.A.).
Wolbachia strain identification was carried out by the amplification of Wolbachia surface protein (WSP) (Baldo et al. 2006). WSP typing was completed in 41 individuals (Suppl. Table S1). After sequencing, we identified the strains in the Wolbachia MLST database (http://pubmlst.org/wolbachia/).
The relative abundance of Wolbachia was quantified using real-time quantitative PCR (RT-QPCR) technology. Wolbachia copy number was determined by measuring 16S rRNA gene level using the W-Spec primers of Werren and Windsor (2000). To calculate the relative abundance of Wolbachia, the copy number of the host-specific histone 3 (H3) gene was measured from the same DNA extract (using H3 primers from Talavera et al. (2013)). Measurements were made with a Quantstudio 12K Flex instrument (Thermo Fisher Scientific, Waltham, MA, USA). The 10-μl reaction mixture contained 5 μl of SYBR Green I Master mix (Roche Life Science, Penzberg, Germany), 0.2 μl of 10 μM primer mixture of forward and reverse primers and 4.8 μl of DNA extract (10 ng/μl). Run profile was as follows: 95 °C for 10 min, followed by 40 cycles of amplification (95 °C for 15 s, 60 °C for 30 s). Specificity of the used primer pairs was checked by the melting curve analysis of the amplified products.
The differences in Wolbachia quantity were tested between the two forms and we also completed pairwise comparisons by sampling sites. As the dataset does not follow normal distribution, we performed a Kolmogorov-Smirnov test using PAST 2.17 (Hammer et al. 2001).
Morphometric analyses
Only males were used for the morphometric survey (altogether 104 specimens, see Suppl. Table S1). The preparation of male external genitalia was performed following the procedure described in Bereczki et al. (2014). Genital slides were digitised using an Olympus camera on a Wild Heerbrugg M420 Microscope. Since we found few reliable landmarks on valvae, we recorded a close curve on it which is basically a series of equally distributed semi-landmarks around the outline of the shape (Fig. 6a) using TpsDig 2.3.1 (Rholf 2015). For the analysis of the outlines, elliptic Fourier analysis (Giardina and Kuhl 1977; Kuhl and Giardina 1982) was used. The algorithm fits Fourier series on x- and y-coordinates as functions of the curvilinear abscissa using PAST 2.17 (Hammer et al. 2001). The Fourier coefficients were analysed in the R computing environment (R Core Team 2018).
Measurement error was calculated by the following formula: ME = Swithin2/(Swithin2 + Samong2) × 100 (Lessells and Boag 1987) where Swithin2 is the within-measurement component of variance and Samong2 is the among-measurement component. Variance components were calculated using ANOVA (Bailey and Byrnes 1990; Yezerinac et al. 1992) in the R computing environment (R Core Team 2018).
Principal component analyses (PCA) were applied on the Fourier coefficients to reduce the number of variables. The scores of the PC axes, which could explain more than 1% of the total variance, were used in a canonical variate analysis (CVA) and a multivariate analysis of variance (MANOVA) (see Dapporto et al. (2009) for more details). Jack-knifed grouping and Wilks’ lambda were applied to quantify the level of separation between groups. Wilks’ lambda measures the discriminatory power of the model. Its value ranges from 0 (perfect discrimination) to 1 (no discrimination). All morphometric analyses were carried out using PAST 2.17 (Hammer et al. 2001) just like the calculation of genital sizes following the method of Kuhl and Giardina (1982). The size difference between the two forms were analysed by nested ANOVA using the ‘nlme’ package in the R computing environment (Pinheiro et al. 2017) where the localities of samples were used as random factor (R Core Team 2018).
Host ant studies
To gather information on possible differences in host ant use of spring and summer arion, we surveyed the potential host ants in those four localities where P. arion had been sampled (Fig. 1). The habitats of the two types of P. arion were partly separated on two of these sites (Korlát hill and Perkupa), while their occurrence completely overlapped in the other two localities (Szin and Zabanyik).
Ten pitfall traps per habitat (altogether 60 traps) were set up to examine the Myrmica species composition. The traps were placed close to one of the food plants (Thymus sp. or Origanum vulgare). In this way, Myrmica falling into the traps could putatively find the dropped P. arion caterpillars and may be considered as potential host ant species. Traps were operated for a week.
Additionally, the habitat was scanned for Myrmica nests. Nests found were carefully opened and five individuals were collected from each of them which were preserved in ethanol for identification and Wolbachia screening. Nests were also checked for P. arion specimens. After excavation, the ground and vegetation were restored as close to the original conditions as possible.
Myrmica species data from the pitfall traps and from the nest searching were combined to a single dataset as follows: species data were counted as presence-absence data per pitfall trap and per nest and then they were summed in each habitat patch. In this way, we could get a picture of how many traps caught the given species and also how many nests of it were found. The collected Myrmica data was visualised on a single map by geo-referenced pie charts.
For barcode-based species identification and Wolbachia screening, DNA was extracted from the whole body of the Myrmica specimens collected from traps and the excavated nests following the protocol of Bereczki et al. (2014). We sequenced the mitochondrial cytochrome c oxidase subunit I gene (COI) using commercial service provider Macrogen. Inc. (Seoul, South Korea). This gene was amplified by modified universal primer pairs HybLCO and HybHCO of Wahlberg et al. (2008) following the amplification protocol presented in Tóth et al. (2014). After the revision of sequences, we identified the specimens at species level using the COI dataset from Jansen et al. (2010). Wolbachia screening and strain identification in Myrmica were carried out in a similar way as in the case of P. arion.
Results
Microsatellite studies
The Micro-Checker analysis showed signs of null alleles in only three cases (at the locus Macari22 in the spring arion sample of Korlát hill, at Macari19 in the summer type from the same locality and at Macari16 in the spring arion sample of Perkupa). Since we did not detect systematic evidence for null alleles at any of the studied microsatellite loci, the whole dataset was used for the further analyses.
Microsatellite variability proved to be generally very high (with 122 alleles in total) without remarkable difference between the spring and summer arion (Table 1). We experienced significant heterozygote deficiency in three samples (see FIS values in Table 1).
Genetic differentiation between the two types of P. arion was pronounced in all of the analyses. The NJ tree based on pairwise FST values showed clear separation between the two phenological forms (Fig. 2). FST values per locus indicated low or moderate (Balloux and Lugon-Moulin 2002), but significant differences between the syntopic sample pairs at 2–4 loci (Table 2).
Unrooted neighbour joining tree based on a pairwise FST matrix. The abbreviations are the same as in Suppl. Table S1. The letter ‘T’ in the sample codes refers to spring arion, the letter ‘N’ indicates summer arion
According to the hierarchical AMOVA, the separation between the two types of P. arion was also evident (see FCTphenology in Table 3). In contrast, we did not detect significant differentiation between the samples when grouping them according to their geographical origin (see FCTlocality in Table 3).
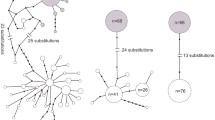
The most probable number of genetically differentiated groups (K) proved to be two in the Structure analysis (Fig. 3). The two clusters corresponded with the phenology of samples, i.e. the spring and the summer arion clearly differed in this analysis as well. Nevertheless, we also detected low gene flow between them since there are spring arion individuals with summer-type genetic background and vice versa. In parallel with this, the number of effective migrants (Nm) between the syntopic sample pairs was 1–2 individuals (Fig. 3) which also corresponds to low gene flow between them (Waples and Gaggiotti 2006).
Results of the Bayesian-clustering Structure analysis based on 9 microsatellite loci. Nm– the number of effective migrants between the syntopic sample pairs using private alleles. The abbreviations are the same as in Suppl. Table S1. The letter ‘T’ in the sample codes refers to spring arion, the letter ‘N’ indicates summer arion
The gene flow was weaker between the spring and summer arion than between the populations in each type (Fig. 4). The number of effective migrants (Nm) was higher in the summer than in the spring form.
The number of effective migrants (Nm) between populations. Only those connections are shown where the number of migrants is greater than two. Thicker arrows indicate higher number of migrants. The abbreviations are the same as in Suppl. Table S1. The letter ‘T’ in the sample codes refers to spring arion, the letter ‘N’ indicates summer arion
Wolbachia screening
The prevalence of Wolbachia was 98.6%. While all summer-type individuals proved to be infected, we could not detect Wolbachia in two spring-type specimens (see Suppl. Table S1). All infected samples harboured a single strain (allele No. 685). The PCR products from Wolbachia screening showed a systematic trend as only faint bands were observed in the spring-type specimens referring to the low abundance of the bacterium (Fig. 5a). On the contrary, strong bands were present in the analysed summer arion referring to high Wolbachia abundance. When we quantified this difference by the RT-Q-PCR method, we detected significantly smaller amounts of Wolbachia in the spring arion than in the summer type comparing the two forms to each other or carrying out pairwise comparisons by sampling sites (Fig. 5b). The summer type contained 29 times more Wolbachia than the spring form considering the median of the Wolbachia quantity in each type of P. arion.
Differences in Wolbachia infestation between the spring and the summer type of P. arion: a differences between the spring and the summer-type specimens from Korlát hill in the Wolbachia quantity on 1% agarose gel; b box-plots showing the difference in infestation levels as measured by RT-Q-PCR between the spring and summer arion in four syntopic population pairs from the Aggtelek Karst region (Hungary). The abbreviations are the same as in Suppl. Table S1. The letter ‘T’ in the sample codes refers to spring arion, the letter ‘N’ indicates summer arion
Morphometric analyses
The measurement error was less than 10% in all cases. Differences in external genital structure between syntopic sample pairs of P. arion were confirmed. The spring and summer type differed from each other significantly (Wilks’ λ = 0.611, P < 0.001). The group centroids of the two phenological forms were clearly separated (Fig. 6b). However, only 74% of the individuals were correctly classified by a cross-validated method. The proportion of the correct classification was 78% in the spring and 70% in the summer type of P. arion. We also detected significant size differences between the two forms (nested ANOVA: fore wing: F = 50.17, df = 1, P < 0.001) (Fig. 6c).
Morphometric studies: a morphometric measurements – close curve on valva of male genitalia; b the results of morphometric analyses: the CVA scatterplots represent the group centroids of the samples; c box plots indicating the distribution of the centroid size. The abbreviations are the same as in Suppl. Table S1. The letter ‘T’ in the sample codes refers to spring arion, the letter ‘N’ indicates summer arion
Host ant studies
Altogether, 247 Myrmica specimens were found on the sampling sites, out of which 195 were collected from 39 nests and 52 were caught by 23 pitfall traps (Suppl. Table 2a). In total, four Myrmica species were identified (Fig. 7). The most common species found on nearly all sites was M. sabuleti Meinert, 1861, which was captured in the largest number in drier habitats (Zabanyik and Korlát hill). In contrast, M. scabrinodis Nylander, 1846, was not found in these drier localities but it was seen in more humid habitats (Perkupa and Szin). M. schencki Viereck, 1903, was recorded in several habitats including the drier Korlát hill and the more humid Perkupa and Szin. M. specioides Bondroit, 1918, was found in very low numbers in two habitat patches: Perkupa where only summer arion was caught and the habitat patch of the spring arion on Korlát hill. This species was found in high number only in Szin where the most common M. sabuleti did not occur.
A total of 151 Myrmica specimens (61%) were infected with Wolbachia (Suppl. Table 2a). The infestation rate was very similar in the case of excavated nests and pitfall traps (62% and 58%, respectively). In most cases, all workers from the same nest were infected or uninfected, although certain nests contained both infected and uninfected specimens (Suppl. Table 2a). All individuals harboured the same Wolbachia strain (the allele No. 59). Myrmica species were different regarding the infestation level (Suppl. Table 2b). M. specioides and M. sabuleti were highly infected (100% and 86%, respectively), while M. scabrinodis and M. schencki had low infection rates (11% and 2%, respectively). However, we have to note that our dataset involved only few individuals of M. specioides and M. schencki.
We found only a single P. arion prepupa (summer type) in a M. specioides nest in Szin. Both the prepupa and all ant workers from this nest (nest ID is 12A, see in Suppl. Table 2a) were infected with Wolbachia but not with the same strain.
Discussion
In this paper, we investigated the nature and causes of difference between the two phenological forms of P. arion which are good candidates for sympatric speciation via host race formation. For this reason, we studied whether these forms meet the criteria of host races according to Drès and Mallet (2002).
-
(i)
Sympatric occurrence
Sympatric speciation requires spatial co-occurrence under the biogeographical definition (Fitzpatrick et al. 2008). The existence of the two forms of P. arion is known from several European countries, e.g. France (Dupont 2010), Italy (Patricelli et al. 2013) or Poland (Sielezniew, pers. comm.). Before our investigations, however, only sporadic data had been available on the coexistence of the two phenological forms in the same habitat, e.g. in Romania (Vizauer, pers. comm.), Russia (Kuznetsov, pers. comm.) and a single locality of Hungary (Varga, pers. comm.). Owing to the intense systematic field mapping, we discovered several sites in the northern part of Hungary, such as the Aggtelek Karst region, where the two arion types occurred syntopically. These populations provide clear evidence of the spatial co-occurrence of the spring and the summer arion in this part of the species range giving an opportunity for initial panmixia which is the prerequisite of sympatric speciation according to the population genetic definition (Mallet et al. 2009).
-
(ii)
Host fidelity
P. arion depends on the dual presence of the specific initial food plant and host ant species. Food plants are selected for oviposition by females on the basis of their bud phenology, i.e. they prefer to lay their eggs on slightly immature buds, thereby ensuring that her brood has enough food for the next two weeks before ants are adopting them (Patricelli et al. 2011). Based on previous botanical surveys (Virók et al. 2016), several different Thymus species occur in P. arion habitats of the studied sites. For example, Thymus pulegoides L. and T. pannonicus All. flower more or less in the flight period of the spring arion. At this time, Origanum vulgare has only vegetative parts as it starts to flower only in July. Therefore, it is not available for the ovipositing females of spring arion. That is, on our sampling sites the phenology of the host plants is usually distinct which is followed by the flight period of P. arion, i.e. there are two more or less separated flight peaks adjusted to the flowering peaks of the food plants. This is what makes the coexistence of the different phenological forms of P. arion possible. The time slot between the appearances of the two arion types varies year by year together with the degree of the overlap between the flowering periods of different host plants (Tóth & Bereczki, pers. obs.). This overlapping phenology, however, hinders the two arion forms to separate entirely. Where there is only one flowering peak of the food plant(s), only a single arion form could occur, for example, if only early flowering Thymus are present in the habitat. That means, the fidelity to a particular host is primarily expressed as the adaptation to the phenology of food plants.
This kind of temporal adaptation to food plant phenology is well exemplified in the two forms of Phengaris alcon ([Denis & Schiffermüller], 1775) exploiting different initial food plants (Gentiana cruciata L. vs. G. pneumonanthe L.) and host ant species accompanied by differential habitat use and phenology. Although their food plants usually grow in different habitats, they co-occur in a few sites where the flowering time of the two Gentiana species is usually separated temporally, but it varies year by year and overlapping phenology occasionally occurs. In such cases, butterflies lay their eggs on both food plants. A previous study on the different forms of P. alcon in the same habitat (Răscruci, Transylvania) has shown that the degree of genetic differentiation between them fluctuated year by year as FST values decreased more than by half from 2007 to 2011 (Bereczki et al. 2018). These dynamics could be the result of varying phenology driven by weather conditions of each year. Larger overlap in the flight period of the two forms could result in a higher level of gene flow.
The host ant specificity of P. arion is less well-known than that of other European Phengaris species (Hayes 2015; Settele et al. 2005; Tartally et al. 2019), because it is very difficult to find P. arion caterpillars in Myrmica nests (Sielezniew et al. 2010a, b). Only two host ant data are known from the Carpathian Basin (Tartally et al. 2017): a spring arion pupa was recorded in a M. scabrinodis nest and a summer arion prepupa with M. specioides. The latter one was recorded in the sampling area of the present study. That means, we do not have enough information on the possible difference in host ant use between the spring and summer arion but we can outline some hypothesis on the potential host ants in general. A strong P. arion population cannot be maintained by a rare Myrmica species with few nests in the habitat. Therefore, the main host or hosts must have reasonably high nest density. Not surprisingly, we detected varying Myrmica species composition on our sampling sites as the studied P. arion populations occurred in different habitat types and the different Myrmica species have distinct ecological needs (e.g. Elmes et al. (1998)). It is also evident that if the potential host ant species are completely different between the habitats, the host ants actually used are also likely to be different. Thus, our data provide a strong indication for the use of multiple host ants across the study area similarly to the Polish populations (Sielezniew et al. 2010a, b; Sielezniew and Stankiewicz 2008).
-
(iii)
Genetic differentiation, (iv) gene flow and (v) spatial and temporal replicability
Previous studies investigating the genetic differentiation between the two arion forms based on mitochondrial and nuclear sequences as well as allozymes have emphasised the need of the application of more variable genetic markers (Bereczki et al. 2011, 2014, 2015). In our study, we applied highly variable microsatellites and we could detect clear differentiation between the two arion forms for the first time. Pairwise FST values demonstrated significant genetic differences between the syntopic sample pairs at more than one locus. The genetic differentiation was higher between the two forms of P. arion than between the locations in each form as reflected by the FST values based NJ tree. Although the number of effective migrants showed only low gene flow between the studied sites, the hierarchical AMOVA could not detect significant differentiation between the localities. These results call our attention that the question of spatial replicability should be also tested in larger geographical context, where gene flow between the populations is more restricted.
The genetic difference has been also confirmed by male genitalia morphometrics, i.e. the spring and summer arion differed from each other significantly based on external genital traits. Good agreement has been revealed between the outcome of the molecular studies and that of male genitalia morphometrics in numerous other analyses (Cesaroni et al. 1989, 1994; Garnier et al. 2004). Moreover, strong phylogenetic signal was detected in the shape of valva in the genus Phengaris (Bereczki et al. 2017). This suggests that selective pressures controlling genital structures are relatively homogeneous across taxa and the patterns of divergence in genital morphology may reflect overall genetic divergence rather than differential adaptive responses. Consequently, the quantitative traits of male genitalia may be good estimators of the overall divergence among populations and closely related species.
Nevertheless, the two forms exchange genes as it was indicated by the Structure analysis and the number of effective migrants. The level of gene flow probably depends on the phenology of the different food plants changing year by year which affects the probability of the meeting of the spring- and summer-type individuals. Moreover, the different microclimate could result in slight shifts in phenology between the different habitats which also influences the length of the time period when the two forms could meet. The number of effective migrants proved to be higher in the summer than in the spring form which is probably due to the longer flight period of the summer arion.
Knowing the level of gene flow between the two arion forms it is not surprising that both of them bear the same Wolbachia strain which was also found on a larger geographical scale in the Carpathian Basin (Bereczki et al. 2015) as well as in Poland and Italy (Patricelli et al. 2013). The infestation pattern, i.e. the nearly 100% prevalence of a single strain is presumably related to the vertical transmission of Wolbachia in this host. In parallel, we detected the presence of another strain in the infected Myrmica specimens, even in the M. specioides nest where P. arion prepupa was found. That means, we could not demonstrate horizontal transfer between P. arion and its ant hosts. Instead, we found high maternal transmission rate in these ant species as it is the case in P. arion. Vertically transmitted endosymbionts are generally considered to be evolved towards mutualism because their evolutionary fate is closely linked to that of their hosts (Forsine and Fine 1975; Yamamura 1993; Zug and Hammerstein 2015). Nevertheless, the phenotype of these Wolbachia strains has remained unknown. Anyway, this infestation pattern, i.e. the presence of a single strain in the two arion forms suggests that Wolbachia does not have any effect on the evolutionary processes in P. arion.
At the same time, the quantity of this intracellular bacterium is significantly different between the two arion forms which is probably due to environmental factors, as it could not be linked to the genetic background (microsatellite cluster) but to the flight time.
Several studies have demonstrated that temperature affects the Wolbachia density within the host cells although the outcome of these surveys is controversial. The individuals of the wasp Leptopilina heterotoma reared at 26 °C harboured approximately twice as many Wolbachia than males which developed at 18 °C (Mouton et al. 2006), while the eggs of Drosophila bifasciata from females maintained at 18 °C harboured more bacteria than eggs from females maintained at 26 °C (Hurst et al. 2000). Similarly, the density of Wolbachia in embryos was lower after exposure to 25 °C than in individuals exposed to 19 °C in D. simulans (Clancy and Hoffmann 1998). These surveys clearly show that the response of Wolbachia density to environmental conditions results from complex genotype-by-genotype-by-environment interactions (Thomas and Blanford 2003), i.e. the effect of temperature will act both directly on the two partners and on their interaction. Since the phenology of the two arion forms are partly distinct they may be subject to various environmental factors which could affect the abundance of Wolbachia in their cells. Nevertheless, further studies are needed to reveal the cause of the difference in Wolbachia quantity between the spring and the summer type of P. arion.
Conclusions
Our results show that the two phenological forms of P. arion seem to meet all criteria of host plant races, although spatial replicability should be tested on a larger geographical scale. The sympatric occurrence of the spring and the summer arion is made possible by the adaptation to the distinct phenology of the host plants. Negative selection acts against the intermediate individuals which are on the wing in the inappropriate time frame. That is, disruptive selection affects and produces bimodal distributions of phenotypes. However, the phenology of food plants is not entirely distinct and fluctuates year by year. Therefore, the two forms can exchange genes occasionally depending on the length of the time slot when they can meet with each other. That is, the reproductive isolation is incomplete thus the existence of the two arion forms may represent only an incipient stage of sympatric speciation. It is also clear that Wolbachia is likely not a driver of sympatric speciation in this case. The evolutionary processes in P. arion, however, cannot be fully understood without further knowledge of its phylogeographical pattern.
Data accessibility
Wolbachia WSP sequence information is available in Wolbachia MLST database.
References
Bailey, R. C., & Byrnes, J. (1990). A new, old method for assessing measurement error in both univariate and multivariate morphometric studies. Systematic Zoology, 39, 124.
Baldo, L., Dunning Hotopp, J. C., Jolley, K. A., Bordenstein, S. R., Biber, S. A., Choudhury, R. R., Hayashi, C., Maiden, M. C. J., Tettelin, H., & Werren, J. H. (2006). Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Applied and Environmental Microbiology, 72, 7098–7110.
Balloux, F., & Lugon-Moulin, N. (2002). The estimation of population differentiation with microsatellite markers. Molecular Ecology, 11, 155–165.
Barton, N. H., & Slatkin, M. (1986). A quasi-equilibrium theory of the distribution of rare alleles in a subdivided population. Heredity, 56, 409–415.
Bereczki, J., Tóth, J. P., Tóth, A., Bátori, E., Pecsenye, K., & Varga, Z. (2011). The genetic structure of phenologically differentiated Large Blue (Maculinea arion) populations (Lepidoptera: Lycaenidae) in the Carpathian Basin. European Journal of Entomology, 108, 519–527.
Bereczki, J., Tóth, J. P., Sramkó, G., & Varga, Z. (2014). Multilevel studies on the two phenological forms of Large Blue (Maculinea arion) (Lepidoptera: Lycaenidae). Journal of Zoological Systematics and Evolutionary Research, 52, 32–43.
Bereczki, J., Rácz, R., Varga, Z., & Tóth, J. P. (2015). Controversial patterns of Wolbachia infestation in the social parasitic Maculinea butterflies (Lepidoptera: Lycaenidae). Organisms, Diversity and Evolution, 15, 591–607.
Bereczki, J., Varga, Z., & Tóth, J. P. (2017). Maculinea or Phengaris? New insights from genitalia morphometry. Zoologischer Anzeiger, 270, 57–59.
Bereczki, J., Pecsenye, K., Varga, Z., Tartally, A., & Tóth, J. P. (2018). Maculinea rebeli (Hirschke) – a phantom or reality? Novel contribution to a long-standing debate over the taxonomic status of an enigmatic Lycaenidae butterfly. Systematic Entomology, 43, 166–182.
Berlocher, S. H., & Feder, J. L. (2002). Sympatric speciation in phytophagous insects: moving beyond controversy? Annual Review of Entomology, 47, 773–815.
Bolnick, D. I., & Fitzpatrick, B. M. (2007). Sympatric speciation: models and empirical evidence. Annual Review of Ecology, Evolution, and Systematics, 38, 459–487.
Bourtzis, K., Nirgianaki, A., Markakis, G., & Savakis, C. (1996). Wolbachia infection and cytoplasmic incompatibility in Drosophila species. Genetics, 144, 1063–1073.
Boyle, L., O’Neill, S. L., Robertson, H. M., & Karr, T. L. (1993). Interspecific and intraspecific horizontal transfer of Wolbachia in Drosophila. Science, 260, 1796–1799.
Breeuwer, J. A., & Werren, J. H. (1993). Cytoplasmic incompatibility and bacterial density in Nasonia vitripennis. Genetics, 135, 565–574.
Bush, G. L. (1969). Sympatric host race formation and speciation in frugivorous flies of the genus Rhagoletis (Diptera, Tephritidae). Evolution, 23, 237–251.
Calvitti, M., Marini, F., Desiderio, A., Puggioli, A., & Moretti, R. (2015). Wolbachia density and cytoplasmic incompatibility in Aedes albopictus: concerns with using artificial Wolbachia infection as a vector suppression tool. PLoS One, 10, e0121813.
Cesaroni, D., Allegrucci, G., Angelici, M. C., Racheli, T., & Sbordoni, V. (1989). Allozymic and morphometric analysis of populations in the Zygaena purpuralis complex (Lepidoptera, Zygaenidae). Biological Journal of the Linnean Society, 36, 271–280.
Cesaroni, D., Lucarelli, M., Allori, P., Russo, F., & Sbordoni, V. (1994). Patterns of evolution and multidimensional systematics in graylings (Lepidoptera: Hipparchia). Biological Journal of the Linnean Society, 52, 101–119.
Clancy, D. J., & Hoffmann, A. A. (1998). Environmental effects on cytoplasmic incompatibility and bacterial load in Wolbachia-infected Drosophila simulans. Entomologia Experimentalis et Applicata, 86, 13–24.
Coyne, J. A. (2007). Sympatric speciation. Current Biology, 17, R787–R788.
Dapporto, L., Bruschini, C., Baracchi, D., Cini, A., Gayubo, S. F., González, J. A., & Dennis, R. L. H. (2009). Phylogeography and counter-intuitive inferences in island biogeography: evidence from morphometric markers in the mobile butterfly Maniola jurtina (Linnaeus) (Lepidoptera, Nymphalidae). Biological Journal of the Linnean Society, 98, 677–692.
Dieckmann, U., & Doebeli, M. (1999). On the origin of species by sympatric speciation. Nature, 400, 354–357.
Drès, M., & Mallet, J. (2002). Host races in plant-feeding insects and their importance in sympatric speciation. Philosophical transactions of the Royal Society of London. Series B, Biological sciences, 357, 471–492.
Dupont, P. (2010). Plant National d’ Actions (PNA) en faveur des Maculinea. Auvergne.
Earl, D. A., & Von Holdt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4, 359–361.
Elmes, G. W., Thomas, J. A., Wardlaw, J. C., Hochberg, M. E., Clarke, R. T., & Simcox, D. J. (1998). The ecology of Myrmica ants in relation to the conservation of Maculinea butterflies. Journal of Insect Conservation, 2, 67–78.
Evanno, G., Regnaut, S., & Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology, 14, 2611–2620.
Excoffier, L., & Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564–567.
Feder, J. L. (1998). The apple maggot fly, Rhagoletis pomonella: flies in the face of conventional wisdom about speciation? In D. J. Howard & S. H. Berlocher (Eds.), Endless forms: species and speciation (pp. 130–144). Oxford: Oxford University Press.
Filchak, K. E., Roethele, J. B., & Feder, J. L. (2000). Natural selection and sympatric divergence in the apple maggot Rhagoletis pomonella. Nature, 407, 739–742.
Fitzpatrick, B. M., Fordyce, J. A., & Gavrilets, S. (2008). What, if anything, is sympatric speciation? Journal of Evolutionary Biology, 21, 1452–1459.
Fitzpatrick, B. M., Fordyce, J. A., & Gavrilets, S. (2009). Pattern, process and geographic modes of speciation. Journal of Evolutionary Biology, 22, 2342–2347.
Forsine, P. E. M., & Fine, P. E. M. (1975). Vectors and vertical transmission: an epidemiologic perspective. Annals of the New York Academy of Sciences, 266, 173–194.
Futuyma, D. J., & Mayer, G. C. (1980). Non-allopatric speciation in animals. Systematic Biology, 29, 254–271.
Garnier, S., Magniez-Jannin, F., Rasplus, J.-Y., & Alibert, P. (2004). When morphometry meets genetics: inferring the phylogeography of Carabus solieri using Fourier analyses of pronotum and male genitalia. Journal of Evolutionary Biology, 18, 269–280.
Gavrilets, S. (2003). Perspective: models of speciation: what have we learned in 40 years? Evolution, 57, 2197–2215.
Giardina, C. R., & Kuhl, F. P. (1977). Accuracy of curve approximation by harmonically related vectors with elliptical loci. Computer Graphics and Image Processing, 6, 277–285.
Goudet, J. (1995). FSTAT (Version 1.2): A Computer Program to Calculate F-statistics. Journal of Heredity, 86, 485–486.
Goudet, J. (2005). HIERFSTAT, a package for r to compute and test hierarchical F-statistics. Molecular Ecology Notes, 5, 184–186.
Hammer, Ø., Harper, D. A. T., & Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Paleontologica Electronica, 4, 9.
Hayes, M. P. (2015). The biology and ecology of the large blue butterfly Phengaris (Maculinea) arion: a review. Journal of Insect Conservation, 19, 1037–1051.
Hoffmann, A. A., & Turelli, M. (1997). Cytoplasmic incompatibility in insects. Oxford: Oxford University Press.
Hoy, M. A., & Jeyaprakash, A. (2005). Microbial diversity in the predatory mite Metaseiulus occidentalis (Acari: Phytoseiidae) and its prey, Tetranychus urticae (Acari: Tetranychidae). Biological Control, 32, 427–441.
Hurst, G. D. D., Hurst, L. D., & Majerus, M. E. N. (1992). Selfish genes move sideways. Nature, 356, 659–660.
Hurst, G. D. D., Johnson, A. P., Hinrich, J. G. v. d. S., Fuyama, Y., & Savakis, C. (2000). Male-killing Wolbachia in Drosophila: a temperature-sensitive trait with a threshold bacterial density. Genetics, 156, 699–709.
Ikeda, T., Ishikawa, H., & Sasaki, T. (2003). Infection density of Wolbachia and level of cytoplasmic incompatibility in the Mediterranean flour moth, Ephestia kuehniella. Journal of Invertebrate Pathology, 84, 1–5.
Jansen, G., Savolainen, R., & Vepsäläinen, K. (2010). Phylogeny, divergence-time estimation, biogeography and social parasite–host relationships of the Holarctic ant genus Myrmica (Hymenoptera: Formicidae). Molecular Phylogenetics and Evolution, 56, 294–304.
Kittayapong, P., Jamnongluk, W., Thipaksorn, A., Milne, J. R., & Sindhusake, C. (2003). Wolbachia infection complexity among insects in the tropical rice-field community. Molecular Ecology, 12, 1049–1060.
Kuhl, F. P., & Giardina, C. R. (1982). Elliptic Fourier features of a closed contour. Computer Graphics and Image Processing, 18, 236–258.
Le Clec’h, W., Chevalier, F. D., Genty, L., Bertaux, J., Bouchon, D., & Sicard, M. (2013). Cannibalism and predation as paths for horizontal passage of Wolbachia between terrestrial isopods. PLoS One, 8, e60232.
Lessells, C. M., & Boag, P. T. (1987). Unrepeatable repeatabilities: a common mistake. The Auk, 104, 116–121.
Mallet, J., Meyer, A., Nosil, P., & Feder, J. L. (2009). Space, sympatry and speciation. Journal of Evolutionary Biology, 22, 2332–2341.
Mouton, L., Henri, H., Bouletreau, M., & Vavre, F. (2006). Effect of temperature on Wolbachia density and impact on cytoplasmic incompatibility. Parasitology, 132, 49–56.
Nosil, P. (2012). Ecological speciation. Oxford: Oxford University Press.
Nowicki, P., Settele, J., Thomas, J. A., & Woyciechowski, M. (2005). A review of population structure of Maculinea butterflies. In J. Settele, E. Kühn, & J. A. Thomas (Eds.), Studies on the Ecology and Conservation of Butterflies in Europe (pp. 144–149). Sofia – Moscow: Pensoft Publishers.
Osváth-Ferencz, M., Bonelli, S., Nowicki, P., Peregovits, L., Rákosy, L., Sielezniew, M., et al. (2017). Population demography of the endangered large blue butterfly Maculinea arion in Europe. Journal of Insect Conservation, 21, 411–422.
Paradis, E., Claude, J., & Strimmer, K. (2004). APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics, 20, 289–290.
Patricelli, D., Barbero, F., La Morgia, V., Casacci, L. P., Witek, M., Balletto, E., & Bonelli, S. (2011). To lay or not to lay: oviposition of Maculinea arion in relation to Myrmica ant presence and host plant phenology. Animal Behaviour, 82, 791–799.
Patricelli, D., Sielezniew, M., Ponikwicka-Tyszko, D., Ratkiewicz, M., Bonelli, S., Barbero, F., et al. (2013). Contrasting genetic structure of rear edge and continuous range populations of a parasitic butterfly infected by Wolbachia. BMC Evolutionary Biology, 13, 14.
Pinheiro, J. C., Bates, D., DebRoy, S., & Sarkar, D. (2017). NLME: Linear and Nonlinear Mixed Effects Models. R Package Version 3.1-129.
Pritchard, J. K., Stephens, M., & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959.
R Core Team. (2018). R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
Rácz, R., Bereczki, J., Sramkó, G., Kosztolányi, A., Tóth, J. P., Póliska, S., et al. (2015). Isolation and characterisation of 15 microsatellite loci from Lethrus apterus (Coleoptera: Geotrupidae). Annales Zoologici Fennici, 52, 45–50.
Raymond, M. (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity, 86, 248–249.
Rholf, F. J. (2015). The tps series of software. Hystrix, 26, 1–4.
Rousset, F. (2008). GENEPOP’007: a complete re-implementation of the genepop software for Windows and Linux. Molecular Ecology Resources, 8, 103–106.
Settele, J., Kühn, E., & Thomas, J. A. (2005). Species ecology along a European gradient: Maculinea butterflies as a model. In J. Settele, E. Kühn, & J. A. Thomas (Eds.), Studies on the Ecology and Conservation of Butterflies in Europe (p. 289). Sofia – Moscow: Pensoft Publishers.
Sielezniew, M., & Rutkowski, R. (2012). Population isolation rather than ecological variation explains the genetic structure of endangered myrmecophilous butterfly Phengaris (=Maculinea) arion. Journal of Insect Conservation, 16, 39–50.
Sielezniew, M., & Stankiewicz, A. M. (2008). Myrmica sabuleti (Hymenoptera: Formicidae) not necessary for the survival of the population of Phengaris (Maculinea) arion (Lepidoptera: Lycaenidae) in eastern Poland: Lower host-ant specificity or evidence for geographical variation of an endangered social parasite? European Journal of Entomology, 105, 637–641.
Sielezniew, M., Dziekańska, I., & Stankiewicz-Fiedurek, A. M. (2010a). Multiple host-ant use by the predatory social parasite Phengaris (=Maculinea) arion (Lepidoptera, Lycaenidae). Journal of Insect Conservation, 14, 141–149.
Sielezniew, M., Patricelli, D., Dziekańska, I., Barbero, F., Bonelli, S., Casacci, L. P., et al. (2010b). The first record of Myrmica lonae (Hymenoptera: Formicidae) as a host of the socially parasitic Large Blue butterfly Phengaris (Maculinea)* arion (Lepidoptera: Lycaenidae). Sociobiology, 56, 465–475.
Sielezniew, M., Patricelli, D., Rutkowski, R., Witek, M., Bonelli, S., & Buś, M. M. (2015). Population genetics of the endangered obligatorily myrmecophilous butterfly Phengaris (= Maculinea ) arion in two areas of its European range. Insect Conservation and Diversity, 8, 505–516.
Smouse, P. E., Whitehead, M. R., & Peakall, R. (2015). An informational diversity framework, illustrated with sexually deceptive orchids in early stages of speciation. Molecular Ecology Resources, 15, 1375–1384.
Talavera, G., Lukhtanov, V. A., Pierce, N. E., & Vila, R. (2013). Establishing criteria for higher-level classification using molecular data: the systematics of Polyommatus blue butterflies (Lepidoptera, Lycaenidae). Cladistics, 29, 166–192.
Tartally, A., Tóth, J. P., Váradi, A., & Bereczki, J. (2017). First data on the host ant usage of Large Blue from the Carpathian Basin. Sociobiology, 64, 122–124.
Tartally, A., Thomas, J. A., Anton, C., Balletto, E., Barbero, F., Bonelli, S., Bräu, M., Casacci, L. P., Csősz, S., Czekes, Z., Dolek, M., Dziekańska, I., Elmes, G., Fürst, M. A., Glinka, U., Hochberg, M. E., Höttinger, H., Hula, V., Maes, D., Munguira, M. L., Musche, M., Nielsen, P. S., Nowicki, P., Oliveira, P. S., Peregovits, L., Ritter, S., Schlick-Steiner, B. C., Settele, J., Sielezniew, M., Simcox, D. J., Stankiewicz, A. M., Steiner, F. M., Švitra, G., Ugelvig, L. V., Van Dyck, H., Varga, Z., Witek, M., Woyciechowski, M., Wynhoff, I., & Nash, D. R. (2019). Patterns of host use by brood parasitic Maculinea butterflies across Europe. Philosophical Transactions of the Royal Society, B: Biological Sciences, 374, 20180202.
Tauber, C. A., & Tauber, M. J. (1989). Symaptric speciation in insects: perception and perspective. In D. Otte & J. Endler (Eds.), Speciation and its Consequences (pp. 307–344). Sunderland: Sinauer Press.
Thomas, M. B., & Blanford, S. (2003). Thermal biology in insect-parasite interactions. Trends in Ecology & Evolution, 18, 344–350.
Thomas, J. A., Elmes, G. W., Wardlaw, J. C., & Woyciechowski, M. (1989). Host specificity among Maculinea butterflies in Myrmica ant nests. Oecologia, 79, 452–457.
Tóth, J. P., Bereczki, J., Varga, Z., Rota, J., Sramkó, G., & Wahlberg, N. (2014). Relationships within the Melitaea phoebe species group (Lepidoptera: Nymphalidae): new insights from molecular and morphometric information. Systematic Entomology, 39, 749–757.
Turelli, M., Barton, N. H., & Coyne, J. A. (2001). Theory and speciation. Trends in Ecology & Evolution, 16, 330–343.
Turner, G. F., & Burrows, M. T. (1995). A model of sympatric speciation by sexual selection. Proceedings of the Royal Society of London. Series B: Biological Sciences, 260, 287–292.
Ugelvig, L. V., Nielsen, P. S., Boomsma, J. J., & Nash, D. R. (2011). Reconstructing eight decades of genetic variation in an isolated Danish population of the large blue butterfly Maculinea arion. BMC Evolutionary Biology, 11, 201.
Ugelvig, L. V., Andersen, A., Boomsma, J. J., & Nash, D. R. (2012). Dispersal and gene flow in the rare, parasitic Large Blue butterfly Maculinea arion. Molecular Ecology, 21, 3224–3236.
Van Meer, M. M. M., Witteveldt, J., & Stouthamer, R. (1999). Phylogeny of the arthropod endosymbiont Wolbachia based on the wsp gene. Insect Molecular Biology, 8, 399–408.
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P. M., & Shipley, P. (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes, 4, 535–538.
Veneti, Z., Clark, M. E., Zabalou, S., Karr, T. L., Savakis, C., & Bourtzis, K. (2003). Cytoplasmic incompatibility and sperm cyst infection in different Drosophila-Wolbachia associations. Genetics, 164, 545–552.
Via, S. (2001). Sympatric speciation in animals: the ugly duckling grows up. Trends in Ecology & Evolution, 16, 381–390.
Virók, V. Farkas, R., Farkas, T., Suvada, R., & Vojtkó, A. (2016). A Gömör–Tornai-karszt flórája Enumeráció [The phytogeography of Gömör–Torna Karst]. Jósvafő, ANP füzetek XIV. (p. 922).
Wahlberg, N., Wheat, C. W., & Knowles, L. L. (2008). Genomic outposts serve the phylogenomic pioneers: designing novel nuclear markers for genomic DNA extractions of Lepidoptera. Systematic Biology, 57, 231–242.
Waples, R. S., & Gaggiotti, O. (2006). What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Molecular Ecology, 15, 1419–1439.
Weir, B. S. (1996). Genetic data analysis II: methods for discrete population genetic data. Sunderland: Sinauer Associates.
Werren, J. H. (1997). Biology of Wolbachia. Annual Review of Entomology, 42, 587–609.
Werren, J. H., & Windsor, D. M. (2000). Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proceedings of the Biological Sciences, 267, 1277–1285.
Werren, J. H., Zhang, W., & Guo, L. R. (1995). Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proceedings of the Biological Sciences, 261, 55–63.
Werren, J. H., Baldo, L., & Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nature Reviews Microbiology, 6, 741–751.
Wright, S. (1978). Variability within and among natural populations. In Evolution and the genetics of populations (p. 590). Chicago: University of Chicago Press.
Yamamura, N. (1993). Vertical transmission and evolution of mutualism from parasitism. Theoretical Population Biology, 44, 95–109.
Yezerinac, S. M., Lougheed, S. C., & Handford, P. (1992). Measurement error and morphometric studies: statistical power and observer experience. Systematic Biology, 41, 471–482.
Zeisset, I., Damm Als, T., Settele, J., & Boomsma, J. J. (2005). Microsatellite markers for the large blue butterflies Maculinea nausithous and Maculinea alcon (Lepidoptera: Lycaenidae) and their amplification in other Maculinea species. Molecular Ecology Notes, 5, 165–168.
Zhou, W., Rousset, F., & O’Neil, S. (1998). Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proceedings of the Biological Sciences, 265, 509–515.
Zug, R., & Hammerstein, P. (2015). Bad guys turned nice? A critical assessment of Wolbachia mutualisms in arthropod hosts. Biological Reviews, 90, 89–111.
Acknowledgements
This work was financed by the Hungarian Scientific Research Fund (OTKA K109223). JB was supported by János Bolyai Scholarship of the Hungarian Academy of Sciences.
Funding
Open access funding provided by University of Debrecen (DE). The technical assistance of V. Mester in the molecular work is very much respected. We are grateful for the advice of K. Pecsenye in statistical analyses and for the help of A. Tartally in the ant studies. We thank the work of M. Tuschek in the Molecular Taxonomy Laboratory of the Hungarian Natural History Museum (Budapest, Hungary). The support of the Nature Conservation Authorities of Hungary is also greatly appreciated.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(PDF 580 kb).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bereczki, J., Póliska, S., Váradi, A. et al. Incipient sympatric speciation via host race formation in Phengaris arion (Lepidoptera: Lycaenidae). Org Divers Evol 20, 63–76 (2020). https://doi.org/10.1007/s13127-019-00418-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-019-00418-y