Abstract
Acute myeloid leukemia (AML) with partial tandem duplication of histone-lysine N-methyltransferase 2A (KMT2A-PTD) is a subtype of AML and is associated with adverse survival, yet the molecular pathogenesis of KMT2A-PTD is not fully understood. DNA methyltransferase 3A (DNMT3A) is mutated in various myeloid neoplasms including AML, especially at the Arg882. Recently, it has been found that DNMT3A mutations frequently coexisted with KMT2A-PTD and are associated with inferior outcomes. We aimed to understand the biological role of DNMT3A mutation in KMT2A-PTD-positive cells. Herein, we found that overexpression of DNMT3A mutants (MT) in KMT2A-PTD-positive EOL-1 cells augmented cell proliferation and clonogenicity. Serial colony replating assays indicated that DNMT3A-MT increased the self-renewal ability of Kmt2a-PTD-expressing mouse bone marrow cells with immature morphology. At 10 months post bone marrow transplantation, mice with the combined Kmt2a-PTD and DNMT3A-MT showed hepatosplenomegaly and leukocytosis with a shorter latency compared to control and DNMT3A-wild-type. Gene expression microarray analyses of bone marrow samples from human AML with KMT2A-PTD/DNMT3A-MT showed a stem cell signature and myeloid hematopoietic lineage with dysregulation of HOXB gene expression. In addition, human bone marrow AML cells carrying KMT2A-PTD/DNMT3A-MT showed abnormal growth and augmented self-renewal activity in primary cell culture. The present study provides information underlying the pathogenic role of DNMT3A-MT with KMT2A-PTD in proliferating advantage with augmentation of self-renewal activity in human leukemia, which may help to better understand the disease and to design better therapy for AML patients with these mutations.
Similar content being viewed by others
Introduction
Acute myeloid leukemia (AML) is a heterogeneous malignant hematopoietic disorder, which is classified by distinct morphologies, cytogenetics and molecular subtypes1. The histone-lysine N-methyltransferase 2A (KMT2A) gene, located on chromosome (11q23), has been found in more than 100 different fusion partners2. The SET domain of KMT2A has histone methyltransferase activity that specifically methylates lysine 4 on histone H3 (H3K4), a modification typically associated with transcriptionally active regions of chromatin3,4,5. Apart from translocation the KMT2A-PTD is an in-frame duplication of KMT2A exons (e.g. e9/e3 and e11/e3) in a 5′ to 3′ direction and produces a gain-of-function elongated protein6,7,8. Our previous study found that 6% of de novo AML patients had KMT2A-PTD mutation, which was associated with poor outcomes9. A Kmt2aPTD/WT mice model was reported not to develop leukemia but characterized by a proliferative advantage, abnormal self-renewal capability, and blockage of myeloid differentiation in hematopoietic stem/progenitor cells10,11.
Because KMT2A-PTD alone does not generate leukemia, the acquisition of other cooperating mutations is required for leukemia transformation. Previously, using a large cohort of 98 de novo AML patients with KMT2A-PTD, we reported that 90.8% of patients had at least one additional gene mutation including FLT3-ITD (44.9%), DNMT3A (32.7%), RUNX1 (23.5%) and TET2 (18.4%) mutations12. We observed not only a high frequency of coexistence of DNMT3A mutations with KMT2A-PTD but also a poor outcome in patients carrying both mutations12. However, the biological functions of DNMT3A mutation in KMT2A-PTD-positive leukemia cells have not been studied before.
Results
Addition of DNMT3A-MT in KMT2A-PTD-positive EOL-1 cells augmented cell proliferation and clonogenicity
To elucidate the functional role of DNMT3A-WT/MT in KMT2A-PTD-positive leukemia cells, we stably expressed DNMT3A-wild-type (WT), DNMT3A-R882C/H (DNMT3A-MT) and empty vector (EV) control in EOL-1 cells (Fig. 1a). EOL-1 cell line was the reported KMT2A-PTD-positive human acute myeloid (eosinophilic) cells13. We assayed cell proliferation, clonogenicity and self-renewal activity of transduced cells. The results showed that cell proliferation, colony formation, and self-renewal activity were significantly higher in EOL-1 cells carrying DNMT3A-MT compared to WT or control (Fig. 1b−d). To check the drug sensitivity, EOL-1 cells were cultured in the presence or absence of various concentrations of all-trans retinoic acid (ATRA) and suberanilohydroxamic acid (SAHA) for 3 days (Supplemental Fig. S1a). To understand the potency of ATRA and SAHA in endogenously expressed KMT2A-WT cells, we used U937 cells (Supplemental Fig. S1b). U937 cell line was one of the myeloid cell lines that expressed WT-KMT2A. We found that cell viability of EOL-1 and U937 cells in the presence of ATRA or SAHA was dissimilar and dependent on different cell context manners. To test the sensitivity of ATRA and SAHA of transduced EOL-1 cells, we cultured DNMT3A-WT/MT-stably expressing cells in the presence of 200 nM ATRA, 600 nM SAHA and the combination of 100 nM ATRA with 500 nM SAHA for 3 days. All drugs effectively reduced cell proliferation at selective doses; reduction of cell proliferation of DNMT3A-MT-expressing cells was less than those of WT or control cells (Fig. 1e). Moreover, it was found that the combination of ATRA and SAHA was more effective compared to the single drug (Fig. 1e). To assess the role of DNMT3A mutants in the differentiation of KMT2A-PTD-positive EOL-1 cells, we incubated transduced EOL-1 cells with ATRA, Na-byturate, and SAHA. Transduced EOL-1 cells treated with ATRA or Na-butyrate induced morphological change with modest differentiation; however, very little effect was observed in EOL-1 cells treated with 500 nM SAHA (Supplemental Fig. S2a, b). With the treatment of 100 nM ATRA, Liu’s reagents stained smears showed a similar morphological feature of transformed EOL-1 cells (Supplemental Fig. S2c). However, EOL-1 cells transduced with DNMT3A-R882H mutant had a lower number but not significantly different expression of CD11b positive cells (Supplemental Fig. S2d). Flow cytometry data showed that CD11b expression was decreased in EOL-1 cells transduced with DNMT3A-MT compared to WT or EV, although there were no major morphological differences after 500 μM Na-butyrate treatment on transduced EOL-1 cells (Supplemental Fig. S2e). In contrast, transformed EOL-1 control cells treated with DMSO alone had no effect on CD11b expression (Supplemental Fig. S2f).
a Immunoblot of DNMT3A overexpressed with WT and mutant-DNMT3A in KMT2A-PTD positive EOL-1 cells. b Growth curves of EOL-1 cells stably transduced with WT- and mutant-DNMT3A. Representative results from three independent replicates are shown. c, d Colony-formation ability in methylcellulose containing medium after stable overexpression of WT; and mutant-DNMT3A in EOL-1 cells (original magnification: ×100). Colonies of more than 50 cells were scored on day 10 of cultures. e Cell viability of transformed EOL-1 cells in the presence of 200 nM ATRA, 600 nM SAHA and the combination of 100 nM ATRA with 500 nM SAHA at 72 h. Error bars represent ± s.d. of the mean of duplicate cultures and each experiment repeated at least three times. *P < 0.05, **P < 0.03, ***P < 0.01, either compared with the control or as indicated in figures. Two-sided Student’s t test was used to calculate the P value.
Primary human KMT2A-PTD/DNMT3A mutants bone marrow cell (BMC) exhibited hyperproliferation, clonogenicity and self-renewal activity
Primary AML cells from four patients (AML#1, AML#2, AML#3 and AML#4) with KMT2A-PTD/DNMT3A-WT and two patients (AML#5 and AML#6) with KMT2A-PTD/DNMT3A-MT were cultured in six-well plate. The characteristics of six AML patients were presented in Supplemental Table S1. To explore the biological effect, we checked cell morphology, assayed cell proliferation, clonogenicity and self-renewal activity of primary human AML cells (Fig. 2a−e). Clonogenicity and self-renewal ability in methylcellulose medium were enhanced in primary AML cells with KMT2A-PTD/DNMT3A-MT compared with KMT2A-PTD/DNMT3A-WT cells (Fig. 2b, c). Results from the colony-forming unit (CFU) assays assessed the type of colonies, granulocyte (CFU-G), macrophage (CFU-M), and granulocyte with macrophage (CFU-GM) (Fig. 2d). Primary AML cell proliferation was significantly increased with more immature cells in KMT2A-PTD/DNMT3A-MT compared to KMT2A-PTD/DNMT3A-WT cells (Fig. 2a, e). Viability of primary AML cells was reduced in the presence of ATRA or SAHA; however, AML cells with KMT2A-PTD/DNMT3A-WT were more sensitive to those drugs when compared with KMT2A-PTD/DNMT3A-MT AML cells (Fig. 2f). Conversely, flow cytometry data showed that primary AML cells with KMT2A-PTD/DNMT3A-MT expressed lower Annexin-V and propidium iodide (PI)-positive cells indicating apoptosis reduction and more viable cells relative to KMT2A-PTD/DNMT3A-WT cells (Fig. 2g, h). It was noted that primary BM cells from one patient (AML#5) had expressed FLT3-ITD with KMT2A-PTD/DNMT3A-MT. Interestingly, in the context of cell proliferation, clonogenicity, and drug-induced cell death, there was no much difference between two human BMC harboring KMT2A-PTD/DNMT3A-MT with FLT3-ITD (AML#5) or without FLT3-ITD (AML#6).
a Representative cell morphology with Liu’s reagents stained smears (original magnification: ×400) of primary human KMT2A-PTD AML cells with DNMT3A-WT/MT. DNMT3A-WT: AML#1, AML#2, AML#3, AML#4; DNMT3A-MT: AML#5, AML#6. b, c Colony-forming potentials and self-renewal activity of KMT2A-PTD/DNMT3A-WT/MT AML bone marrow cells in human MethoCult H4535 medium-enriched without erythropoietin (EPO), representative images of colonies in the second-round replating were shown. Colonies of more than 30 cells were scored using an inverted microscope on day 10 of cultures (b), original magnification: ×100. Columns represent the number of serially replated colonies (c). d Proportion of CFU-colonies of first-round plating. e Primary AML cell proliferation at day 3 (3d) and day 6 (6d) in a liquid culture medium containing 20% FBS and 20% conditional medium. f Cell viability of primary AML cells harboring KMT2A-PTD/DNMT3A-WT/MT in the presence of 200 nM ATRA, 600 nM SAHA incubated for 72 h. Each experiment repeated two times and error bars represent ±s.d. of duplicate cultures. g Representative flow cytometry data to determine the Annexin-V and propidium iodide (PI)-positive primary AML cells. h Primary BM cells were cultured in RPMI medium containing 20% FBS, 20% conditional medium with antibiotic for 3 days and the percentage of Annexin V and PI-positive cells were analyses by flow cytometry. Data are presented as means ± s.d. (n = 3). Two-sided Student’s t test was used to calculate the P value and compared between KMT2A/DNMT3A-WT and KMT2A/DNMT3A-MT groups, n.s. not significant.
DNMT3A mutants deregulate hematopoietic stem cells (HSCs) activation, proliferation and RNA modifying-associated genes in primary AML cells with KMT2A-PTD
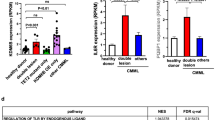
To understand the mechanism underlying the biological effect of DNMT3A mutants in KMT2A-PTD-positive leukemia, we examined the patient samples harboring both KMT2A-PTD and DNMT3A-WT/MT by using global gene expression microarray analysis. Previously we had analyzed 25 AML samples of which 15 were KMT2A-PTD and 10 were KMT2A-translocation14. From these datasets, we analyzed seven samples, four KMT2A-PTD with DNMT3A-WT and three KMT2A-PTD with DNMT3A-R882 mutants (accession no. GSE15013). The characteristics of patients and disease subtypes are shown in Supplemental Table S2. Gene expression microarray data revealed that 762 and 209 genes were upregulated and downregulated, respectively, more than 2-folds in KMT2A-PTD AML with DNMT3A mutations compared to DNMT3A-WT samples (Supplemental Datasets S1, S2). We found that a group of genes associated with HSC activation, positive regulation of cell proliferation and myeloid differentiation was upregulated in DNMT3A-MT samples (Fig. 3a). HOXB genes were upregulated in KMT2A-PTD AML with DNMT3A mutations. Upregulated genes in KMT2A-PTD AML samples with DNMT3A-MT compared to KMT2A-PTD with DNMT3A-WT were used for Gene Ontology (GO) classification and enrichment analysis. Indeed, several upregulated gene sets were enriched in different cellular and molecular processes including lymphocyte proliferation, neutrophil activation, T-cell differentiation, regulation of angiogenesis (Fig. 3b). In addition, analysis based on the Hematopoietic Fingerprints Database15 revealed 83 genes were upregulated in KMT2A-PTD AML samples with DNMT3A mutation compared to KMT2A-PTD with DNMT3A-WT (Supplemental Table S3) samples. Gene set enrichment analyses (GSEA) of gene expression data revealed that the mutation of DNMT3A with KMT2A-PTD significantly altered the expression profile compared to KMT2A-PTD with DNMT3A-WT (Fig. 3c−f). GSEA indicated that the gene set induced by DNMT3A-WT with KMT2A-PTD positively correlated with those of enriched in mRNA processing, RNA splicing and RNA methylation activated gene sets, and negatively correlated with those enriched in platelet activation when compared with the gene sets induced by DNMT3A mutant with KMT2A-PTD AML (Fig. 3c−f).
a Heatmap representation of HSC activation, positive regulation of cell proliferation, HOXB gene expression identified as being differentially expressed in human primary AML cells harboring KMT2A-PTD with DNMT3A-WT and mutants. Red indicates upregulated genes compared to WT (green). b GO analyses of upregulated genes in DNMT3A-MT with KMT2A-PTD AML cells showing a series of genes enriched in different cellular and molecular processes including cell proliferation, angiogenesis, and regulation of apoptosis. c−f GSEA determining specific gene sets or pathways that are positively or negatively regulated by DNMT3A mutants with KMT2A-PTD. Compared with KMT2A-PTD/DNMT3A-MT AML (right side), KMT2A-PTD/DNMT3A-WT (left side) negatively correlated with gene sets downregulated in platelet activation (c), positively correlated with those of mRNA processing (d), RNA splicing (e) and RNA methylation (f). The enrichment scores (ES), normalized enrichment scores (NES) and P values were shown in figures.
DNMT3A-MT upregulates HOXB gene expression in KMT2A-PTD-positive EOL-1 and primary AML cells
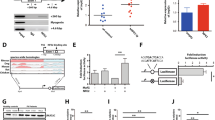
From gene expression microarray data analyses, we found that several genes including the HOXB cluster were upregulated in KMT2A-PTD AML with DNMT3A mutations compared to DNMT3A-WT. A portion of the differentially expressed genes was validated in EOL-1 cells stably expressed-DNMT3A-WT/MT (Supplemental Fig. S3a). Among them, antiapoptotic genes BCL2A1 and MCL1 that act as a key driver of survival in AML were also upregulated in mutant cells16,17. Moreover, we found that HOXB cluster genes including HOXB2, HOXB3, HOXB5, and HOXB8 were upregulated in EOL-1 cells expressing DNMT3A-MT when compared with EV or DNMT3A-WT (Fig. 4a). However, expression of HOXA cluster genes including HOXA5, HOXA7, HOXA9, and HOXA10 was not changed in mutant cells compared to either EV or WT cells (Supplemental Fig. S3b). Immunoblot data showed that EOL-1 cells transduced with DNMT3A-MT had increased but not a significantly different expression of H3K4me3 and significantly decreased expression of H4Ac compared to WT cells (Fig. 4b, c). We then asked whether DNMT3A mutation affected the status of H4 acetylation at the locus of HOXB cluster genes. ChIP assays were performed with antibodies against H4Ac. ChIP-qPCR for H4Ac in EOL-1 cells carrying DNMT3A-R882H mutation and DNMT3A-WT revealed a reduction of H4Ac enrichment at the HOXB promoter regions with R882H mutation compared to DNMT3A-WT (Fig. 4d). Similarly, qRT-PCR analyses showed that primary BM cells harboring both KMT2A-PTD and DNMT3A-MT upregulated HOXB (B2, B3, B4, and B5) expression compared to cells with KMT2A-PTD/DNMT3A-WT (Fig. 4e). Immunoblot data showed that DNMT3A expression was downregulated in KMT2A-PTD/DNMT3A-MT AML cells; conversely, the expression of BCL2 and CDK1 was upregulated in similar cells compared to KMT2A-PTD/DNMT3A-WT AML cells (Supplemental Fig. S4). However, the expressions of H3K4me2, H3K4me3, H3, H3Ac, and H4Ac was comparable between the two groups (Supplemental Fig. S4).
a HOXB expression in EOL-1 cells transduced with DNMT3A-WT/MT by quantitative RT-PCR analyses showing the same patterns observed in gene expression microarray analysis of patient samples with KMT2A-PTD/DNMT3A-WT and mutants. Data are expressed as mean ± s.d. of three independent experiments. *P < 0.05, ***P < 0.005 compared to EV. Two-sided Student’s t test was used to calculate the P value. b Immunoblot data showing H3K4me3 and H4Ac protein levels increased and decreased respectively in EOL-1 cells expressing DNMT3A-MT. β-Actin was used as a control for equal loading. c Quantitation of indicated proteins in transduced EOL-1 cells. Error bars presented as mean ± s.d. of three independent experiments. *P < 0.02, **P < 0.006; n.s. not significant. Two-sided Student’s t test was used to calculate the P value. d Levels of H4Ac at the promoters of HOXB genes in DNMT3A-WT and DNMT3A-R882H-expressing EOL-1 cells as detected by ChIP-qPCR. The relative amounts of immunoprecipitated DNA were depicted as a percentage of input DNA. Error bars presented as mean ± s.d. of two independent experiments. *P < 0.05, ***P < 0.005. Two-sided Student’s t test was used to calculate the P value. e Relative expression levels of HOXB genes were examined by quantitative RT-PCR in BM cells derived from normal control (n = 3) and from primary AML cells with KMT2A-PTD/DNMT3A-WT/MT. The values were normalized by GAPDH mRNA levels and expression was shown as relative to normal BM cells (average of 3). Experiment was repeated twice and data are expressed as mean ± s.d. of two experiments.
DNMT3A-MT induces modifications of genomic methylation patterns in KMT2A-PTD-positive EOL-1 cells
Next, we analyzed the DNA methylation status of EOL-1 cells transduced with WT-, mutant DNMT3A, and control to assess whether DNMT3A mutant altered gene expression profiles were due to their changes of methyltransferase activity. Indeed, both DNA-hypomethylation and hypermethylation features were observed in the specific region throughout the whole genome (Fig. 5a). Overall, R882C mutation was more hypomethylated and less hypermethylated compared to EV or WT-expressing EOL-1 cells (Fig. 5b). Also, the changes in hypo- and hypermethylation patterns were seen in the context of gene structure, namely promoter, gene body, the transcriptional termination region (TTR), and the intergenic region. We found that R882C mutation was more hypomethylated in the intergenic and gene body regions, whereas WT- and control cells were more hypermethylated in those regions (Fig. 5c, d). We then examined the methylation patterns in four regions defined by the distance from the CpG islands18, such as CpG islands, Shore, Shelf, and Open Sea regions. Most of the hypo- and hypermethylation patterns were identified in the Open Sea region (Fig. 5e, f). In the context of gene methylation patterns, we found that the CCL5 gene was differentially methylated in promoter regions and AREG mainly in gene body region (Supplemental Fig. S5a, b) of DNMT3A-R882C EOL-1 cells. To understand the relationship between global DNA methylation and gene expression changes, we examined upregulated genes in KMT2A-PTD AML samples with DNMT3A-MT compared to KMT2A-PTD with DNMT3A-WT (Supplemental Dataset S1). We analyzed the DNA methylation status of those genes in EOL-1 cells expressing DNMT3A-WT and R882C mutation. We observed that 399 genes out of 762 upregulated genes were less methylated (differential β value < −0.3) in EOL-1 cells expressing R882C compared to DNMT3A-WT (Supplemental Dataset S3), indicating the reduction of methyltransferase activity due to DNMT3A mutation. In contrast, 49 genes were more methylated (differential β value > 0.3) in EOL-1 cells expressing R882C compared to DNMT3A-WT (Supplemental Dataset S4a). Whereas genomic features of 29 genes showed both decreased (differential β value < −0.3) and increased (differential β value > 0.3) methylation at different genomic regions in EOL-1 cells expressing R882C compared to DNMT3A-WT (Supplemental Dataset S4b). Moreover, 276 genes of 762 upregulated genes were hypomethylated (β value < 0.25) in EOL-1 cells expressing R882C compared to DNMT3A-WT (Supplemental Dataset S5).
a Distribution of genomic methylation pattern (β value) in the whole genome of EOL-1 cells transduced with EV control, DNMT3A-WT, and DNMT3A-R882C. b Hypo- and hypermethylation probes count obtained from transduced EOL-1 cells shown as bar-diagram. β value < 0.25 and >0.75 considered as hypomethylation and hypermethylation peaks, respectively. c, d The total hypermethylation and hypomethylation probes counted in each region defined by genomic structure shown in bar graph. e, f Methylation patterns in four regions defined by the distance from CpG islands, such as CpG islands, Shore, Shelf, and Open Sea region were shown.
Expression of DNMT3A-MT in Kmt2a-PTD-expressing mouse BMC enhanced clonogenic and self-renewal activity
To determine the effect of DNMT3A-MT on clonogenicity and self-renewal activity in mouse BMC harboring Kmt2a-PTD, we transduced Kmt2a-PTD-positive mouse BMC with DNMT3A-WT/MT and EV, and transgene expressions were checked by qRT-PCR (Fig. 6a). We examined their colony-forming ability (Fig. 6b) and self-renewal activity (Fig. 6c) in M3434 medium and analyses type of colonies, granulocyte, macrophage, and granulocyte with macrophage (Fig. 6d). However, we could not observe erythroid (CFU-E) and burst-forming unit erythroid (BFU-E) colonies. We found that DNMT3A-MT-transduced Kmt2a-PTD-positive BM cells had significantly greater colony-formation ability with relatively immature morphology (Fig. 6c, e, f) compared with either DNMT3A-WT or empty vector control BM cells. Moreover, DNMT3A-MT-transduced cells produced significantly more colonies in the second- and third-round plating compared with DNMT3A-WT-expressing cells (Fig. 6c). Similarly, more dense colonies were seen in second-round plating of the DNMT3A-MT/Kmt2a-PTD BM cells indicating that DNMT3A-MT cells may maintain an immature phenotype longer than DNMT3A-WT cells.
a Lentivirus-mediated transduction of DNMT3A-WT/mutants in Kmt2a-PTD-positive mouse BM cells and DNMT3A mRNA level was detected by quantitative RT-PCR using first colony-forming cells. b, c Colony-forming potentials of Kmt2a-PTD+ mouse BM cells transduced with indicated plasmids in methylcellulose colony-forming media, representative images of colonies in the second-round plating were shown. Colonies at least 50 cells were scored on day 8 of cultures. Colonies were photographed and counted manually, original magnification: ×100. Columns represent the number of serially replated colonies. Error bars represent the mean ± s.d. from duplicated cultures. *P < 0.05, **P < 0.03, ***P < 0.01, compared with the control. d Proportion of CFU-colonies of second-round serial replating were shown. e Cytospin smeared preparations of cultured cells in the first plating colonies assays by Liu’s reagents staining were shown. f Statistical analysis of the numbers of immature cells in the first plating colonies cells. Error bars represent the mean ± s.d. of 4–5 different microscopic fields. **P < 0.01, ***P < 0.001, compared with the control. Two-sided Student’s t test was used to calculate the P value.
The collaboration of DNMT3A-MT with Kmt2a-PTD in the myeloid transformation
To explore the in vivo transformation effect of DNMT3A-WT/MT with KMT2A-PTD, we carried out bone marrow transplantation (BMT) experiments using murine BMC harboring Kmt2a-PTD, lentiviral transduced with DNMT3A-WT/MT, and EV constructs. To determine whether susceptibility to hematologic malignancy was affected by the cooperation of Kmt2a-PTD with DNMT3A-WT/MT, we monitored mice up to 10 months. Mice carrying either empty vector or DNMT3A-WT displayed no hematologic abnormalities. At 10 months post transplantation, 2 and 3 of 5 mice transplanted with Kmt2a-PTD-BM cells expressing DNMT3A-R882C and DNMT3A-R882H/S mutants, respectively, died with marked splenomegaly and hepatomegaly compared with Kmt2a-PTD/EV-transplanted mice (Fig. 7a−c). The survival of DNMT3A-WT/MT and EV control mice are shown in Fig. 7a. In contrast, all of the five mice transplanted with Kmt2a-PTD BM cells expressing DNMT3A-WT constructs were healthy and alive at the same time. Peripheral blood counts showed leukocytosis (Fig. 7d−f and Supplemental Fig. S6a) but there was no anemia or thrombocytopenia in the Kmt2a-PTD/DNMT3A-MT-transplanted mice (Supplemental Fig. S6b, c). Flow cytometric analyses of lineage markers from the bone marrow cells of Kmt2a-PTD/DNMT3A-MT-transplanted mice revealed more Sca-1-, cKit/Sca-1- and CD34-positive cells and lesser number of Mac-1 (CD11b) and CD14-positive cells (Supplemental Fig. S7).
a Kaplan−Meier curve shows the survival of mice transplanted with BM cells harboring Kmt2a and transduced with empty vector (EV, n = 5) control, DNMT3A-WT, DNMT3A-R882C/H/S (n = 5). P values were calculated using a log-rank test. b, c Quantification of hepatomegaly (b) and splenomegaly (c) in mice transduced with EV, DNMT3A-WT, DNMT3A-R882C/H/S were shown (n = 5). d−f Peripheral blood counts of transduced mice were shown (n = 5 for each group). *P < 0.05, **P < 0.01, ***P < 0.005, compared with the DNMT3A-WT group.
Discussion
It is unclear how KMT2A-PTD contributes to AML. Potential mechanisms of how KMT2A-PTD regulates normal hematopoiesis have been reported19,20. Repetitive DNA-binding domain in KMT2A-PTD exhibits transcriptional potential in vitro19 and KMT2A-PTD contributes to the leukemic phenotype in KMT2A-WT AML cells via a recessive gain-of-function mechanism20. Little is known about the mechanism of crosstalk and cooperation between KMT2A-PTD and other gene mutations in the leukemogenesis. We have observed a high frequency of coexistence of DNMT3A mutations with KMT2A-PTD; more importantly, it conferred a very poor outcome in our AML patients12. Moreover, most DNMT3A mutations (67.7%) were in the methyltransferase domain at amino acid R88212. The present results suggested that DNMT3A-R882 mutations with KMT2A-PTD promoted myeloid malignancy transformation in vitro and in vivo.
In accordance with the in vitro cooperation of DNMT3A-MT with KMT2A-PTD, we observed the transduction of DNMT3A-MT in EOL-1 cells, a KMT2A-PTD-positive leukemias cell line, augmented cell growth and clonogenicity, as well as increased self-renewal activity of the transduced-primary murine BMC-expressing Kmt2a-PTD. The EOL-1 cell line endured eosinophilic differentiation upon Na-butyrate treatment and ATRA regulated the expression of CD11b but did not induce mature eosinophil morphology21,22. During the eosinophilic differentiation of EOL-1 cells, the expression of CD11b was increased under differentiating stimuli23,24. In primary human BM cells expressing KMT2A-PTD and DNMT3A-MT showed hyperproliferation and self-renewal clonogenic potential compared to DNMT3A-WT with KMT2A-PTD, suggesting a critical role of mutant DNMT3A in AML patients with KMT2A-PTD. It was reported that ATRA reduced cell growth with the induction of apoptosis of EOL-1 cells22. We found that on the treatment of either ATRA or SAHA, DNMT3A-WT/MT-transduced EOL-1 cells showed inhibition of cell growth, and the combination of both agents was more effective than a single agent. However, ATRA was more potent than SAHA in transduced EOL-1 cells. The growth-inhibitory potency of ATRA or SAHA on EOL-1 cells was higher compared to U937 cells indicating that the mechanism of growth inhibition of these two drugs was different in different cell lines harboring endogenous KMT2A or KMT2A-PTD. Moreover, ATRA and SAHA significantly reduced the proliferation of primary KMT2A-PTD AML cells with DNMT3A-WT/MT. However, these agents were more active in the reduction of cell growth in KMT2A-PTD/DNMT3A-WT AML cells compared to KMT2A-PTD/DNMT3A-MT cells. One possible explanation of this finding was that DNMT3A mutations induced HSC expansion, cooperated with KMT2A-PTD to induce AML and contribute to the resistance to these drugs. Though it might be possible that cooperation of other oncogenic abnormalities with DNMT3A mutation in EOL-1 cells, the co-occurrence of KMT2D-PTD with DNMT3A mutation should play a role at least partly in the leukemogenesis in the current situation as DNMT3A mutations frequently coexist with KMT2A-PTD. Previously, it was demonstrated that the combination of decitabine, a hypomethylating agent and HDAC (histone deacetylase) inhibitor, vorinostat (also known as SAHA) decreased the transcription ratio of the KMT2A-PTD-to-KMT2A-WT alleles and simultaneously induced apoptosis in KMT2A-PTD-positive blasts25. In our animal model, we could not find an acute leukemia transformation in Kmt2a-PTD mice with DNMT3A-MT, but we observed hematologic abnormalities with shorter latency and hepatosplenomegaly compared to control or DNMT3A-WT mice. It has been reported that either KMT2A-PTD or Flt3-ITD do not develop AML; however, the cooperation of Flt3-ITD with Kmt2a-PTD developed acute leukemia in a mouse model26. It was also reported that the Dnmt3a mRNA level was increased in BM from double knock-in (Kmt2a-PTD and Flt3-ITD) mice compared to single knock-in or WT control26. However, we observed that downregulation of the DNMT3A level in KMT2A-PTD/DNMT3A-MT AML primary cells with FLT3-ITD or without FLT3-ITD relative to KMT2A-PTD/DNMT3A-WT, suggesting that DNMT3A-WT/MT status had maintained DNMT3A expression rather than FLT3-ITD status. Previously, Huang et al.27 reported that DNMT3A expression was significantly decreased with DNMT3A-R882 mutations and the reduced expression levels were associated with a lower complete remission rate in the AML patients. It was also reported that Arg882 mutants reduced their DNA methylation activities compared to DNMT3A-WT28. Therefore, lower amounts of DNMT3A protein might reduce enzymatic activities in AML patients with DNMT3A mutations, which regulate crucial gene expression in the pathogenesis of AML in KMT2A-PTD-positive leukemic cells. Recently, we found that overexpression of DNMT3A-R882C/H mutations enhanced clonogenicity, cell proliferation, and growth with impairment of apoptosis in the endogenously KMT2A-WT-expressing U937 and HL-60 cells with different mechanisms29.
To test the transformational ability of DNMT3A mutation in KMT2A-PTD AML, we investigated gene expression profiles of BM cells of KMT2A-PTD with DNMT3A-WT/MTs. Previously, we found that AML with KMT2A-PTD can be distinguished from AML with KMT2A-translocation on the basis of HOXB gene clusters14. Analyses of a subset of the same microarray dataset, we found that AML with KMT2A-PTD/DNMT3A mutations enhanced the HOXB gene expression compared with KMT2A-PTD/DNMT3A-WT which was consistent with our in vitro data in EOL-1 cells expressing DNMT3A-WT/MT. These results demonstrated that the DNMT3A function was important for HOXB gene regulation in human leukemia cells harboring KMT2A-PTD. Similarly, primary BM cells from AML patients with KMT2A-PTD/DNMT3A mutations showed higher HOXB expression compared to KMT2A-PTD/DNMT3A-WT, which indicate a critical role of HOXB in AML with KMT2A-PTD/DNMT3A-MT. We observed that KMT2A-PTD was frequently associated with FLT3-ITD in AML12; however, previously using three large-scale datasets analyses, we reported that upregulation of HOXB in KMT2A-PTD might not be related to FLT3-ITD status14. Gene expression profiling analyses of transduced U937 cells with DNMT3A-MT revealed aberrant expression of several cell-cycle and apoptosis-related genes; however, no differentially expressed HOXB compared to DNMT3-WT-transduced U937 cells have been reported recently (accession number GSE90934)29. It was demonstrated that Kmt2a-PTD induced aberrant Hox expression including Hoxa7, Hoxa9, and Hoxa10 in a mouse model30. In contrast, we could not find the upregulation of HOXA including HOXA9 in our study. We also found that the expression of series of genes associated with HSC activation, myeloid differentiation and cell survival, such as MEIS1, HBEGF, BCL6, and MCL1, was significantly upregulated in patients with KMT2A-PTD/DNMT3A-MT. The transcription factor Meis1 was associated with long-term hematopoiesis and always coactivated with Hoxa7 or Hoxa9 in leukemic mice31,32. In addition to the upregulation of MCL-1, pathways involved in granulocytic/monocytic lineage proliferation, including HOXB2, RAB20, SOCS3, and MEIS1, were activated, and those involved in platelet activation, including PRKCA, PF4, and ITGA2B, were upregulated in KMT2A-PTD AML with DNMT3A mutations. RNA modifications, including RNA methylation, mRNA processing, and RNA splicing, play a critical role in important biological processes33,34. RNA modifications have been associated with embryonic stem cell maintenance and differentiation35,36,37. By using GSEA analyses of primary KMT2A-PTD AML with DNMT3A-WT/MT, we identified several deregulated pathways. Among them, the RNA methylation pathway is critical in AML, as the previous study of other investigators showed that genetic alterations of m6A (N6-methyladenosine) regulatory genes are associated with worse survival and inferior outcome in AML patients38. N6-methyladenosine was the most established and internal modification that occurred in the mRNA which was catalyzed by a methyltransferase complex containing methyltransferase-like 3 (METTL3), RNA methyltransferase-like 1 (RNMTL1) and methyltransferase-like 1 (METTL1), were downregulated, and Wilm’s-tumor-1-associated protein (WTAP)33,39 was upregulated in KMT2A-PTD AML with DNMT3A mutations.
DNMT3A is a DNA methyltransferase that catalyzes DNA methylation. Both KMT2A and DNMT3A regulate normal hematopoiesis. Cancer progression involves a diversity of hypo- and hypermethylated genomic regions, resulting in dramatic alterations in gene expression patterns40. From DNA methylation microarray analyses using transformed EOL-1 cells, we observed that DNMT3A-R882C mutation had both DNA methylation-dependent and -independent effects and exhibited distinct mechanism for leukemogenesis. DNMT3A-R882C-expressing EOL-1 cells were more hypomethylated in the gene body and intergenic regions rather than promoter region compared to DNMT3A-WT-expressing cells. However, there was no much difference in the CpG region. Similar results were observed in U937 cells transduced with DNMT3A-WT/MT (GSE90934)29. The results indicated that DNMT3A-WT/MT but not KMT2A-WT or KMT2A-PTD were responsible for the hypo- or hypermethylation features in cells. We next compared the DNA methylation status and gene expression changes in EOL-1 and primary AML cells, respectively using two different datasets. It was observed that many upregulated genes in KMT2A-PTD AML with DNMT3A-MT were less methylated and some genes were more methylated in EOL-1 cells expressing DNMT3A-R882C mutation compared to DNMT3A-WT, indicating that DNMT3A mutations controlled the gene expression changes by both DNA methylation-dependent and -independent manner in KMT2A-PTD AML. It was observed that a group of upregulated genes associated with HSC activation and positive regulation of cell proliferation, including CD34, HBEGF, STOX2, MEIS1, MCL1, PCNX, AREG, and HLX, less methylated in EOL-1 cells expressing DNMT3A-R882C mutation compared to DNMT3A-WT. Loss of Dnmt3a was shown to upregulate multipotency genes, downregulate differentiation factors, and progressively impair differentiation in HSCs, attributable to both increased and decreased methylation at distinct loci41. The previous study illustrated that DNMT3A mutations with reduced enzymatic activity or aberrant affinity to histone H3 would alter DNA methylation patterns and/or gene expression profiles such as HOXB upregulation due to hypomethylation in AML28.
Up to date, to the best of our knowledge, the present study is the first to determine the biological role of DNMT3A mutation in KMT2A-PTD-positive cells, which might contribute to leukemogenesis in vitro and in vivo. The present study provides additional information underlying the pathogenic role of DNMT3A-MT with KMT2A-PTD in proliferating advantage with stem cell signature, enhanced self-renewal activity and aberrant HOXB expression in human leukemia, which may help to better understand the disease and to design better therapy for AML patients with these mutations.
Materials and methods
Reagents and antibodies
RPMI 1640, FBS and antibiotic-antimitotic were purchased from Thermo Fisher Scientific (Waltham, MA, USA); puromycin (101-58-58-2) from MD Bioscience; human and mouse methocult medium, MethoCult H4535 and MethoCult M3434 respectively from STEMCELL Technologies (Vancouver, BC, Canada); mouse stem cell factor and recombinant mouse IL-3 from R&D (Minneapolis, MN, USA); polybrene (H9268), ATRA (R2625), SAHA (SML0061) and β-Actin (A5441) from Sigma-Aldrich (St. Louis, MO, USA); anti-DNMT3A (#3598) from Cell Signaling Technology (Danvers, MA, USA); ChIP Assay Kit (#17–295), anti-H3K4me2 (#07–030), anti-H3Ac (#17–615), anti-H4Ac (#06866) from Millipore (Temecula, CA, USA); anti-H3K4me3 (ab8580), anti-BCL2 (ab692) and anti-H3 (ab70550) from Abcam (Cambridge, UK); Liu’s reagents A and B (03R011/03R021) from ASK Biotech (Taiwan); trypan blue from Gibco-Life Technologies Corporation (Grand Island, NY, USA) and anti-CDK1 (# GTX20018) from GeneTex (Irvine, CA).
Cell lines, mouse and human primary BM cells
EOL-1 and U937 cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) and 1× antibiotic-antimycotic in a humidified chamber with 5% CO2 atmosphere at 37 °C. EOL-1 cell line was obtained from the laboratory of Dr. Gang Huang, Cincinnati Children’s Hospital Medical Center (Cincinnati, USA) and U937 from our own stocks. Both cell lines were authenticated by cellular morphology and STR analysis at Chang Gung Memorial Hospital (CGMH) (January−February 2017). Primary human BM cells were collected at initial diagnosis according to the CGMH guidelines after getting written consent from patients. The study was approved by the Institutional Board of CGMH. Liquid nitrogen storing (<1 year) cells were cultured in MethoCult H4535 medium for 5 days, then washed with complete RPMI medium and cultured in RPMI medium containing 20% FBS, 20% conditional medium (5637 cells cultured soup) and 1× antibiotic; medium changed every 2 days interval. BM samples having more than 90% of viable cells were selected for biological analyses. Kmt2a-PTD+ mouse BM cells were generated and maintained in the laboratory of Dr. Gang Huang. The development of the Kmt2aPTD/WT mouse has been described previously10,11. Mouse BM cells were maintained by the M3434 MethoCult medium. In each experiment, cells were washed by phosphate-buffered saline (PBS) and subsequently cultured in RPMI 1640 medium supplemented with 20% FBS; 1× antibiotic-antimitotic, 100 ng/ml mouse stem cell factor, and 10 ng/ml recombinant mouse IL-3. For morphological studies, cytospin smears were stained with Liu’s reagents. Digital images were acquired using Olympus (model no. U-TV0.5XC-3) microscope equipped with a digital camera.
Plasmid construction, lentiviral preparation, and infection
The full-length cDNA of human DNMT3A-WT and point mutants, R882C, R882H, and R882S, were constructed as described previously29. Lentiviruses production and infection were performed as described previously42. The infected cells were selected with 2 µg/ml puromycin for 2 weeks to obtain stable clones.
Mice and bone marrow transplantation (BMT) experiment
All animal care and experiments were approved by the Department of Animal Experimentation at CGMH. Female C57BL/6 mice (NARLabs, Taiwan) aged 6−8 weeks were used for the BMT experiments. Mouse BMT was performed as described previously43. Briefly, the murine BM cells harboring Kmt2a-PTD were used for transduction of DMMT3A-WT/MT with lentiviruses by spun inoculation in the presence of 8 μg/ml polybrene. The infection was repeated after 48 h of the first infection and transduced BM cells (1 × 106 cells/mouse) were transplanted into intraperitoneally injected busulfan (a single dose of 30 mg/kg prior to 3 days) mice via tail vein44,45. Mice were monitored every day up to 10 months and moribund mice were euthanized according to the animal house guidelines.
Cell proliferation assay
Viable cells were assessed with either the trypan blue exclusion method or using flow cytometry analyses staining with Annexin V and propidium iodide at different time points. To check the sensitivity of different drugs, 1 × 105/ml EOL-1 and U937 cells were cultured in the presence of ATRA and SAHA at different concentrations for 72 h and single dose of each drug was selected to test the activity in transformed EOL-1 cells. To check cell proliferation assays, 2 × 105/ml human primary cells were cultured in liquid medium for 3 and 6 days. To check the sensitivity of ATRA and SAHA, human primary cells were cultured in the presence of 200 nM ATRA and 600 nM SAHA for 3 days, respectively. The percentage of viable cells was calculated by comparing the number of drug-treated cells to that in the untreated control cells.
Colony formation and self-renewal activity assays
For clonogenic growth assays, transformed stable EOL-1 cells were cultured in 12-well plate at 2 × 103 cells/well in 1% methylcellulose containing RPMI medium supplemented with 10% FBS for 10 days. 5 × 104 transduced mouse BM cells were mixed with 2 ml MethoCult M3434 medium in six-well plate and cultured for 8 days. For human primary BM colony-formation assays, cells were seeded in six-well plates with 2 ml H4535 medium at a density of 5 × 104 cells/well. The photograph was taken by a phase-contrast microscope (Nikon Eclipse TS100, Japan). For serial replating assays, all cells were harvested, washed twice with RPMI medium and counted. A similar number of cells were then replated and the process was repeated four times to check colony formation and self-renewal activity. The results from colony-forming unit (CFU) assays assessed granulocyte (CFU-G; colorless, more dense smaller and round cells), macrophage (CFU-M; colorless, less dense, large and elongated cells), granulocyte with macrophage (CFU-GM; colorless, heterogeneous population of small, round cells and large, elongated cells) and erythroid (CFU-E; red color either very small colonies or BFU-E; clusters containing group of tiny cells in irregular shape).
Flow cytometry analysis
Flow cytometry experimentation was described in a previous publication29. Anti-CD11b PE (#12–0112) was purchased from eBioscience (San Diego, CA) and the apoptosis was detected by Annexin V antibody and propidium iodide staining according to the manufacturer’s instructions (Annexin V-FITC kit, #556547, BD Bioscience, CA).
Isolation of RNA, reverse transcription and quantitative real-time polymerase chain reaction (qRT-PCR)
Methods were described previously in detail29. Briefly, after RNA extraction, samples were reverse-transcribed to cDNA and used for quantitative PCR with iQTM SYBR® Green Supermix (ABI, Thermo Fischer Scientific) according to the manufacturer’s protocol. The sequences of oligonucleotides used for qRT-PCR are listed in Supplementary Table S4.
Immunoblotting analysis
Immunoblot was performed with the following antibodies: anti-DNMT3A, anti-H3K4me2, anti-H3Ac, anti-H4Ac, anti-H3K4me3, anti-BCL2, anti-H3, anti-CDK1, and anti-β-actin. Cell lysates were extracted with RIPA buffer and subjected to SDS-PAGE and Western blot analysis as described previously29. A loading control of the detection of β-actin was included for all immunoblots.
Chromatin immunoprecipitation (ChIP) analysis
The ChIP assays were carried out according to the manufacturer’s (Millipore) protocol. Briefly, 1 × 106 transduced EOL-1 cells were used for each immunoprecipitation. Cells were cross-linked by adding 1% formaldehyde, collected, resuspended in ChIP lysis buffer and sonicated to obtain chromatin fragments in a size between 200 and 1000 base pairs. DNA−protein complexes were then immunoprecipitated with a 5 µl anti-H4Ac antibody. Protein A Agarose/Salmon Sperm DNA beads were used to capture the antibody−chromatin complex. The immunoprecipitated samples and an aliquot of the input were subjected to reverse cross-link. Purified CHIP product was quantified by RT-PCR using SYBR Green on an ABI Prism 7900HT Fast Real-Time PCR system (Thermo Fisher Scientific). Quantitative real-time PCR quantification of ChIP was performed in duplicate using primers (Supplementary Table S5) specific for promoter regions.
DNA methylation microarray analysis
Genome-wide DNA methylation was assessed using the Illumina Infinium Human MethylationEPIC Beadchip (Illumina Inc, CA, USA) according to the manufacturer’s instructions. Briefly, genomic DNA was extracted and purified from all samples using a column-based DNA extraction kit (Qiagen DNeasy Kit) according to the standard method. Five hundred nanograms of DNA of each sample was subjected to bisulfite conversion with the EZ DNA methylation kit (Zymo Research) according to the manufacturer’s instructions. Bisulfite converted DNA was amplified, fragmented and hybridized to Illumina Infinium Human MethylationEPIC Beadchip using standard Illumina protocol. The sample was labeled with Cy5 and Cy3 using Illumina standard protocol. Arrays were imaged using iSCAN using standard recommended Illumina scanner setting. Illumina GenomeStudio software v2010.3 was used to extract the raw signal intensities of each probe (without background correction or normalization). The resulting raw data were normalized (control normalization) and background corrected by the manufacturer to generate methylation β values. Genomic methylation patterns (hypo- or hypermethylation) of different samples were analyzed after the variance filter (STD > 0.2) and raw data (without filter) used for the analyses of a specific gene methylation pattern. β values <0.25 and >0.75 were considered as hypomethylation and hypermethylation peaks, respectively. The data, including all raw genomic-methylation data, have been deposited in the Gene Expression Omnibus (GEO) database with accession number GSE109364.
Statistical analysis
The Kaplan–Meier analysis was used to evaluate mouse survival. Differences in survival were assessed using the log-rank test. All in vitro data represented here are mean ± s.d. as indicated. The significance of the differences between groups was determined using the two-sided Student’s t test. A P value of <0.05 was considered significant for all analyses.
References
Arber, D. A. et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127, 2391–2405 (2016).
Meyer, C. et al. The MLL recombinome of acute leukemias in 2013. Leukemia 27, 2165–2176 (2013).
Milne, T. A. et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10, 1107–1117 (2002).
Tenney, K. & Shilatifard, A. A COMPASS in the voyage of defining the role of trithorax/MLL-containing complexes: linking leukemogensis to covalent modifications of chromatin. J. Cell. Biochem. 95, 429–436 (2005).
Cosgrove, M. S. & Patel, A. Mixed lineage leukemia: a structure-function perspective of the MLL1 protein. FEBS J. 277, 1832–1842 (2010).
Schichman, S. A. et al. ALL-1 tandem duplication in acute myeloid leukemia with a normal karyotype involves homologous recombination between Alu elements. Cancer Res. 54, 4277–4280 (1994).
Zeleznik-Le, N. J., Harden, A. M. & Rowley, J. D. 11q23 translocations split the “AT-hook” cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc. Natl. Acad. Sci. USA 91, 10610–10614 (1994).
Steudel, C. et al. Comparative analysis of MLL partial tandem duplication and FLT3 internal tandem duplication mutations in 956 adult patients with acute myeloid leukemia. Genes Chromosomes Cancer 37, 237251 (2003).
Shih, L. Y. et al. Characterization of fusion partner genes in 114 patients with de novo acute myeloid leukemia and MLL rearrangement. Leukemia 20, 218–223 (2006).
Dorrance, A. M. et al. The Mll partial tandem duplication: differential, tissue-specific activity in the presence or absence of the wild-type allele. Blood 112, 2508–2511 (2008).
Zhang, Y. et al. Stress hematopoiesis reveals abnormal control of self-renewal, lineage bias, and myeloid differentiation in Mll partial tandem duplication (Mll-PTD) hematopoietic stem/progenitor cells. Blood 120, 1118–1129 (2012).
Kao, H. W. et al. High frequency of additional gene mutations in acute myeloid leukemia with MLL partial tandem duplication: DNMT3A mutation is associated with poor prognosis. Oncotarget 6, 33217–33225 (2015).
Drexler, H. G., Quentmeier, H. & MacLeod, R. A. Malignant hematopoietic cell lines: in vitro models for the study of MLL gene alterations. Leukemia 18, 227–232 (2004).
Liu, H. C. et al. Expression of HOXB genes is significantly different in acute myeloid leukemia with a partial tandem duplication of MLL vs. a MLL translocation: a cross-laboratory study. Cancer Genet. 204, 252–259 (2011).
Chambers, S. M. et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell 1, 578–591 (2007).
Powell, J. A. et al. Targeting sphingosine kinase 1 induces MCL1-dependent cell death in acute myeloid leukemia. Blood 129, 771–782 (2017).
Jenal, M. et al. The anti-apoptotic gene BCL2A1 is a novel transcriptional target of PU.1. Leukemia 24, 1073–1076 (2010).
Sandoval, J. et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 6, 692–702 (2011).
Martin, M. E. et al. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer Cell 4, 197–207 (2003).
Whitman, S. P. et al. The MLL partial tandem duplication: evidence for recessive gain-of-function in acute myeloid leukemia identifies a novel patient subgroup for molecular-targeted therapy. Blood 106, 345–352 (2005).
Ishihara, K. et al. Differentiation of eosinophilic leukemia EoL-1 cells into eosinophils induced by histone deacetylase inhibitors. Life Sci. 80, 1213–1220 (2007).
Robert, C. et al. Apoptosis induction by retinoids in eosinophilic leukemia cells: implication of retinoic acid receptor-alpha signaling in all-trans-retinoic acid hypersensitivity. Cancer Res. 66, 6336–6344 (2006).
Jung, E. Y. et al. Effects of cyclic AMP on expression of LFA-1, Mac-1, and VLA-4 and eosinophilic differentiation of a human leukemia cell line, EoL-1. Eur. J. Haematol. 53, 156–162 (1994).
Wong, C. K., Ho, C. Y., Lam, C. W., Zhang, J. P. & Hjelm, N. M. Differentiation of a human eosinophilic leukemic cell line, EoL-1: characterization by the expression of cytokine receptors, adhesion molecules, CD95 and eosinophilic cationic protein (ECP). Immunol. Lett. 68, 317–323 (1999).
Whitman, S. P. et al. DNA hypermethylation and epigenetic silencing of the tumor suppressor gene, SLC5A8, in acute myeloid leukemia with the MLL partial tandem duplication. Blood 112, 2013–2016 (2008).
Zorko, N. A. et al. Mll partial tandem duplication and Flt3 internal tandem duplication in a double knock-in mouse recapitulates features of counterpart human acute myeloid leukemias. Blood 120, 1130–1136 (2012).
Huang, X. et al. Gene expression profiling of the DNMT3A R882 mutation in acute leukemia. Oncol. Lett. 6, 268–274 (2013).
Yan, X. J. et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat. Genet. 43, 309–315 (2011).
Bera, R. et al. Genetic and epigenetic perturbations by DNMT3A-R882 mutants impaired apoptosis through augmentation of PRDX2 in myeloid leukemia cells. Neoplasia 20, 1106–1120 (2018).
Dorrance, A. M. et al. Mll partial tandem duplication induces aberrant Hox expression in vivo via specific epigenetic alterations. J. Clin. Invest. 116, 2707–2716 (2006).
Unnisa, Z. et al. Meis1 preserves hematopoietic stem cells in mice by limiting oxidative stress. Blood 120, 4973–4981 (2012).
Thorsteinsdottir, U., Kroon, E., Jerome, L., Blasi, F. & Sauvageau, G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol. Cell Biol. 21, 224–234 (2001).
Cui, Q. et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18, 2622–2634 (2017).
Li, Z. et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell 31, 127–141 (2017).
Batista, P. J. et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014).
Wang, Y. et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198 (2014).
Geula, S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006 (2015).
Kwok, C. T., Marshall, A. D., Rasko, J. E. & Wong, J. J. Genetic alterations of m(6)A regulators predict poorer survival in acute myeloid leukemia. J. Hematol. Oncol. 10, 39 (2017).
Yue, Y., Liu, J. & He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355 (2015).
Robertson, K. D. DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610 (2005).
Challen, G. A. et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 44, 23–31 (2011).
Tsai, S. C. et al. Biological activities of RUNX1 mutants predict secondary acute leukemia transformation from chronic myelomonocytic leukemia and myelodysplastic syndromes. Clin. Cancer Res. 21, 3541–3551 (2015).
Bersenev, A. et al. Lnk constrains myeloproliferative diseases in mice. J. Clin. Invest. 120, 2058–2069 (2010).
Robert-Richard, E. et al. Human cell engraftment after busulfan or irradiation conditioning of NOD/SCID mice. Haematologica 91, 1384–1387 (2006).
Wilkinson, F. L. et al. Busulfan conditioning enhances engraftment of hematopoietic donor-derived cells in the brain compared with irradiation. Mol. Ther. 21, 868–876 (2013).
Acknowledgements
We thank Mr. Tung-Huei Lin for preparing mice survival curves. This work was supported by grants from the National Health Research Institute, Taiwan (NHRI-EX103-10003NI), the Ministry of Science and Technology, Taiwan (MOST 103-2321-B-182-015, MOST 104-2321-B-182-005), the Ministry of Health and Welfare, Taiwan (MOHW103-TD-B-111-09), and grants from Chang Gung Memorial Hospital, Taipei, Taiwan (OMRPG380031, OMRPG3C0021, and 101-1409B).
Author information
Authors and Affiliations
Contributions
R.B. designed and performed the research, analyzed the data, created the figures, and wrote the manuscript; M.-C.C. performed plasmid construction and technical helped; Y.-J.H. performed flow cytometry analysis; G.H. generated and maintained of Kmt2a-PTD+ mouse BM cells, Y.-S.L performed microarray data analysis and L.-Y.S. planned and implemented the research, provided patients’ samples and clinical data, supervised the experimental work and data analyses, and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bera, R., Chiu, MC., Huang, YJ. et al. DNMT3A mutants provide proliferating advantage with augmentation of self-renewal activity in the pathogenesis of AML in KMT2A-PTD-positive leukemic cells. Oncogenesis 9, 7 (2020). https://doi.org/10.1038/s41389-020-0191-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41389-020-0191-6