Abstract
The metabolism-controlled differentiation of αβ T cells has been well documented; however, the role of a metabolism program in γδ T cell differentiation and function has not been clarified. Here, using CD2-cre; mTORC1 Raptor-f/f, and mTORC2 Rictor-f/f mice (KO mice), we found that mTORC1, but not mTORC2, was required for the proliferation and survival of peripheral γδ T cells, especially Vγ4 γδ T cells. Moreover, mTORC1 was essential for both γδ T1 and γδ Τ17 differentiation, whereas mTORC2 was required for γδ T17, but not for γδ Τ1, differentiation. We further studied the underlying molecular mechanisms and found that depletion of mTORC1 resulted in the increased expression of SOCS1, which in turn suppressed the key transcription factor Eomes, consequentially reducing IFN-γ production. Whereas the reduced glycolysis resulted in impaired γδ Τ17 differentiation in Raptor KO γδ T cells. In contrast, mTORC2 potentiated γδ Τ17 induction by suppressing mitochondrial ROS (mitoROS) production. Consistent with their cytokine production profiles, the Raptor KO γδ T cells lost their anti-tumor function both in vitro and in vivo, whereas both Raptor and Rictor KO mice were resistant to imiquimod (IMQ)-induced psoriasis-like skin pathogenesis. In summary, we identified previously unknown functions of mTORC1 and mTORC2 in γδ T cell differentiation and clarified their divergent roles in mediating the activity of γδ T cells in tumors and autoimmunity.
Similar content being viewed by others
Introduction
γδ T cells are considered innate-like lymphocytes and play important roles in protective immunity against infections and tumors [1,2,3,4,5,6]. Similar to αβ T cells, γδ T cells can produce different arrays of cytokines, such as IFN-γ, IL-4, and IL-17, and are named γδ Τ1, γδ Τ2, and γδ Τ17, respectively.
Our group has extensively studied the molecular mechanisms underlying γδ T cell differentiation, especially IFN-γ production by γδ T cells and their roles in tumor immunity [7,8,9].
Recently, γδ T cell effector fates, especially development of γδ T1 and γδ T17, which are defined by IFN-γ and IL-17 production, respectively, were recognized to be pre-determined in the thymus [10, 11]. It is believed that γδ TCR antigen-naïve cells develop into IL-17 producers, while antigen-experienced cells secrete IFN-γ in the thymus [10]. Another important regulator of γδ T1 vs γδ T17 development is CD27: CD27+ γδ T cells are directed to the γδ T1 fate, whereas CD27- cells differentiate into the γδ T17 fate [11]. Although TCR signaling was found to be required for the maximum differentiation of both γδ T1 and γδ T17 cells [12] and strong TCR signals suppress the γδ T17 fate [13], controversial results have been reported, such that γδ T17 cells were found to originate from SOX13+ DN1d progenitors, which are independent of TCR signaling [14].
In addition to the thymic development theory mentioned above, the extrathymic programming of γδ T1 and γδ T17 cells from peripheral naïve cells has been reported [15, 16]. However, the molecular mechanisms by which γδ T cells are programmed are not well understood.
The mammalian (or mechanistic) target of rapamycin (mTOR) is an important signal transducer that integrates environmental signals, cell metabolism, and differentiation for immune cells [17]. However, the roles of the mTOR complexes in driving γδ T cell differentiation and function remain unclear. A recent study identified mTORC1 directs thymic DN progenitor cells to the αβ lineage [18]. In our previous study, we also found that rapamycin, inhibitor of mTORC1, promoted γδ T cell NKG2D and TNF-α expression [19].
In this study, we found that mTORC1 directs both the γδ T1 and γδ T17 differentiation of peripheral γδ T cells by the mechanisms involving glycolysis. mTORC2 promotes γδ T17 differentiation via inhibiting mitoROS production. γδ T cells from Raptor KO, failed to control tumor growth, whereas both Raptor KO and Rictor KO mice were resistant to psoriasis. The results of this study thus established distinct functions of mTOR complexes in γδ T cell differentiation.
Results
mTORC1 signaling maintains peripheral γδ T cell numbers
The mTOR signaling pathway has been found to be involved in the biological functioning of αβ T cells and other immune cells; however, it has not yet been studied in γδ T cells. We investigated the expression of mTOR, Raptor, and Rictor in lymphoid organs ex vivo, and found that all of them were highly expressed and further induced following activation with different kinetics (Fig. 1a, b). CD71 and CD98, downstream of mTORC1, were found to be expressed higher in CD27+ γδ T cells and Vγ4 γδ T cells (Fig. 1c, d), indicating the involvement of mTORC1 signaling in the functional regulation of γδ T cells.
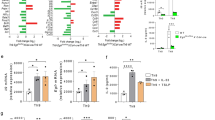
a γδ, CD4 and CD8 T cells were sorted from pooled LNs, spleens, and thymus of WT mice (n = 20). Cells were lysed for Real-time PCR analysis for mTOR, Raptor and Rictor expression. b γδ T cells were sorted from pooled splenocytes of WT mice (n = 18), stimulated with anti-γδ TCR and cells were collected at different time points for Real-time PCR analysis. c, d γδ T cells isolated from pooled splenocytes of WT mice (n = 10) and stimulated with anti-γδ TCR for 48 h and analyzed for CD98 and CD71 expression in CD27−/CD27+ gate (c) and Vγ1 /Vγ4 gate (d). In each plot, the numbers following the gate label (in black or red) represent mean fluorescence intensity (MFI) for the stained marker. e, f Raptor-f/f and Raptor KO mice were analyzed for CD3+ T cell or γδ T cell numbers in spleen ex vivo. Each symbol represents an individual mouse, statistics of 4–5 mice for each group was shown. g Vγ1 and Vγ4 subsets were analyzed for γδ TCR+ CD3+ gate in (f), each symbol represents an individual mouse and statistics for 4–5 mice was shown. h, i Rictor-f/f and Rictor KO mice were analyzed for CD3+ T cell or γδ T cell numbers in spleen ex vivo. Each symbol represents an individual mouse. Statistics of 3–6 mice for each group was shown. j Vγ1 and Vγ4 subsets were analyzed for γδ TCR+ CD3+ gate in (i). Each symbol represents an individual mouse, statistics for 3–6 mice was shown. k, l The expression of Ki-67 was analyzed by FACS gated on γδ T cells from spleen ex vivo. Each symbol represents an individual mouse. Statistics of five mice for each group was shown in (k) and representative plots were shown in (l). m γδ T cells were isolated from Raptor-f/f (n = 5) and Raptor KO (n = 10) mice and activated by anti-γδ TCR antibody for 48 h and apoptosis was analyzed by co-staining of Annexin V and PI. Bim (n) and active caspase-3 (o) were stained for γδ T cells in Raptor-f/f and Raptor KO mice from spleen ex vivo. Each symbol represents an individual mouse, statistics of 3–6 mice for each group was shown. p Strategy of bone marrow chimera for Raptor-f/f and Raptor KO mice. Bone marrow (BM) cells from WT (CD45.1) and Raptor KO (CD45.2) mice were isolated and mixed at a ratio of 1:1 and injected into irradiated TCR δ-/- hosts. Two months after transfer, mice were analyzed. q, r Chimera mice were analyzed for thymus, spleen γδ T cells. Each symbol represents an individual mouse, statistics of 10 recipient mice for each group was shown in (q). Representative FACS plots with gates shown at the top of plots were shown in (r). Data are means ± SEM. ns, not significant; *P < 0.05, **P < 0.01 and ***P < 0.001 (two-tailed unpaired t test). Data are representative of at least three independent experiments. Numbers indicate percentage of cells in quadrants or gates.
To investigate the roles of mTOR complexes in γδ T cell differentiation and function, we used conditional knockout mice in our research. We crossed Raptor-f/f and Rictor-f/f with CD2-cre mice to generate Raptor-f/f; CD2-cre (Raptor KO) and Rictor-f/f; CD2-cre (Rictor KO) mice to clarify the function of mTORC1 or mTORC2, respectively. In these mice, Raptor or Rictor was knocked out in all lymphocyte subsets (αβ and γδ T cells and B cells) [18]. We confirmed the deletion of the Raptor or Rictor protein successful and mTOR downstream signals were weakened in γδ T cells from Raptor KO or Rictor KO mice (Fig. S1A–D). We examined the cell numbers of total T cells as well as γδ T cells in these KO mice. Depletion of Raptor greatly reduced αβ T cell numbers as well as γδ T cell numbers in the spleen (Fig. 1e, f, S1E, S1F). Interestingly, Raptor KO mice exhibited a selective reduction in the percentage of Vγ4 γδ T cells, with normal percentages of Vγ1 γδ T cells (Fig. 1g). On the other hand, Rictor KO mice had reduced αβ T cell numbers (Fig. 1h, S1G, S1H) and normal γδ T cell numbers (Fig. 1i), and the percentage of the Vγ1 subset was slightly increased, with no change in the Vγ4 percentage in γδ T cells (Fig. 1j). To further investigate the underlining mechanisms, we analyzed Ki-67, a proliferation marker, in Raptor KO spleen γδ T cells. Ki-67 expression was significantly reduced in Raptor KO γδ T cells (Fig. 1k, l). Consistently, sorted γδ T cells from Raptor KO mice showed reduced proliferation following either TCR or anti-CD3 activation in in vitro culture (Fig. S2). Furthermore, enhanced apoptosis in Raptor KO γδ T cells was observed following activation (Fig. 1m–o). In contrast, Rictor KO mice had normal numbers of γδ T cells in the spleen (Fig. 1i), with no change in the proliferation rate (Fig. S3A) and the percentage of apoptosis (Fig. S3B). To determine whether the regulation of γδ T cells by mTORC1 was cell intrinsic, WT (CD45.1) and Raptor KO (CD45.2) bone marrow cells were mixed at a ratio of 1:1 and injected into irradiated TCRδ−/− hosts intravenously. The host mice were then analyzed 2 months post-bone marrow transfer (Fig. 1p). Raptor KO cells failed to maintain the number of γδ T cells, especially the Vγ4 subset in the spleen (Fig. 1q, r), indicating that intrinsic mTORC1 signaling was required for the maintenance of peripheral γδ T cell numbers. Together, our results demonstrated that mTORC1, but not mTORC2, signaling was required for the maintenance of γδ T cells, especially the Vγ4 γδ T cell subset, through promoting proliferation and inhibiting apoptosis.
mTORC1 impacts both γδ T1 and γδ T17 differentiation, but mTORC2 impacts only γδ T17 differentiation
The mTOR signaling pathways play essential roles in many aspects of CD4 and CD8 T cells [20,21,22,23]. To determine the effect of mTOR signaling on the functional differentiation of γδ T cells, we analyzed the percentages of γδ T1 and γδ T17 in splenocytes samples ex vivo. γδ T cells from Raptor KO mice showed significant decreases in both IFN-γ+ and IL-17+ production (Fig. 2a, b). In contrast, in Rictor KO mice, only the percentage of γδ T17 cells was reduced, with a normal percentage of γδ T1 cells in the periphery (Fig. 2c, d). To further verify the effects of mTOR signaling on the intrinsic differentiation ability of γδ T cells, naïve γδ T cells were sorted from WT, Raptor or Rictor KO mice and primed under Th1 or Th17 differentiation conditions as described in our previous study [7]. Similar to the cytokine profiles ex vivo, the differentiation of both γδ Τ1 and γδ Τ17 cells were significantly impaired in Raptor KO mice (Fig. 2e–h). However, only γδ T17 differentiation was reduced in Rictor KO cells. The cytokine profiles of the bone marrow chimeric mice were consistent with those of the KO mice (Fig. S4A–D), indicating that the mediation of γδ T cell cytokine regulation by mTORC1 and mTORC2 was cell intrinsic. The reduction in γδ Τ1 differentiation in Raptor KO mice was not due to insufficient proliferation, as indicated by the reduction of γδ T1 in CTVlow cells after culture in vitro (Fig. S4E). Thus, our results clearly demonstrated that mTORC1 impacted both γδ T1 and γδ T17 differentiation but that mTORC2 only impacted γδ T17 differentiation.
a, b Raptor-f/f and Raptor KO mice were sacrificed and γδ T cells were analyzed for IL-17 and IFN-γ expression in spleen ex vivo. Each symbol represents an individual mouse, statistics of 5 mice for each group shown in (a). Representative plots were shown in (b). c, d Rictor-f/f and Rictor KO mice were analyzed as in (a) and statistics of 4–5 mice for each group was shown in (c) and representative plots were shown in (d). e–h Naïve γδ T cells were sorted from pooled splenocytes of WT (n = 15), Raptor KO (n = 17) or Rictor KO (n = 14) mice and cultured under γδ T1 (e, f) or γδ T17 (g, h) -skewing conditions. At day 4, cells were collected for IFN-γ / IL-17 staining. Statistics for triplicates was shown in (e, g) and representative results in (f, h). Data are means ± SEM. ns, not significant; *P < 0.05, **P < 0.01 and ***P < 0.001 (two-tailed unpaired t test). Data are representative of at least three independent experiments. Numbers indicate percentage of cells in quadrants.
mTORC1 mediates γδ T cell IFN-γ production through upregulation of Eomes expression
Since T-bet and Eomes are key transcription factors for controlling IFN-γ production in T cells [9, 24, 25], we analyzed the expression levels of T-bet and Eomes in peripheral γδ T cells from Raptor KO mice ex vivo. Interestingly, the expression level of Eomes was significantly reduced in peripheral γδ T cells from Raptor KO mice, with only a slight reduction in the expression of T-bet (Fig. 3a). In contrast, the expression level of T-bet was significantly reduced in peripheral γδ T cells from Rictor KO mice, and the level of Eomes was not affected (Fig. S5A, S5B). To determine the roles of T-bet and Eomes in γδ T cell IFN-γ production, we crossed CD2-cre mice with T-bet-f/f or Eomes-f/f mice to generate T-bet or Eomes conditional KO mice. We confirmed that the deletion was complete in γδ T cells from these mice (Fig. S6A–D). Indeed, the level of IFN-γ production by γδ T cells from Eomes KO mice was significantly reduced, with no fluctuations in IL-17 secretion (Fig. 3c, d). This trend was especially apparent in the CD27+ γδ T subset (Fig. 3e, f). γδ T cells isolated from T-bet KO mice showed reduced IFN-γ production (Fig. 3g, h), especially in the CD27+ γδ T subset (Fig. 3i, j), indicating a non-redundant role of T-bet in IFN-γ production by γδ T cells, which is consistent with previous findings [9, 26]. These results provide evidence for the role of Eomes in mediating the mTORC1 promotion of IFN-γ production.
a, b Raptor-f/f and Raptor KO mice were sacrificed and γδ T cells were analyzed for T-bet and Eomes expression in spleen ex vivo. Each symbol represents an individual mouse. Statistics of 4–5 mice for each group was shown in (a) and representative staining gated on γδ T cells was shown in (b). c–f Eomes-f/f and Eomes KO mice were analyzed for γδ T cell or CD27+ γδ T cell IL-17 and IFN-γ staining in spleen ex vivo. Each symbol represents an individual mouse. Statistics of 4 mice for each group was shown in (c, e) and representative plots for Eomes-f/f and Eomes KO γδ T cells was shown in (d, f). g–j T-bet-f/f and T-bet KO mice were analyzed for γδ T cell or CD27+ γδ T cell IL-17 and IFN-γ staining in spleen ex vivo. Each symbol represents an individual mouse. Statistics of 6 mice for each group in (g, i) and representative FACS plots were shown in (h, j). Data are means ± SEM. ns, not significant; *P < 0.05, **P < 0.01 and ***P < 0.001(two-tailed unpaired t test). Data are representative of at least two independent experiments. Numbers indicate percentage of cells in quadrants.
mTORC1 promotes Eomes through inhibiting SOCS1 expression
To further investigate the underlying mechanisms by which mTORC1 upregulates Eomes expression, γδ T cells from Raptor KO and Rictor KO mice were sorted for RNAseq analysis. Consistent with the cytokine profiles, γδ T cells from Raptor KO mice showed a broad reduction in both γδ T1 and γδ T17 signature genes, while Rictor KO γδ T cells only showed a decrease in γδ T17-related genes (Fig. 4a). The results indicated that the expression of SOCS1 significantly increased in Raptor KO γδ T cells (Fig. 4a). The results were further validated by analyzing SOCS1 expression by real-time PCR (Fig. 4b). The expression levels of SOCS1 in γδ T cells sorted from Raptor KO mice increased significantly following in vitro activation under γδ T1 condition (Fig. 4b). To test the possibility that Raptor KO-triggered SOCS1 might suppress IFN-γ production in γδ T cells, an siRNA approach was applied. γδ T cells were sorted from Raptor KO mice and activated under γδ T1 conditions in the presence of either siControl (siCtrl) or siSOCS1 for 4 days, and the cells were then restimulated for intracellular cytokine staining. The results indicated that the knockdown of SOCS1 restored IFN-γ production by γδ T cells from Raptor KO mice (Fig. 4c, d). Interestingly, the knockdown of SOCS1 in Raptor KO γδ T cell also significantly increased Eomes expression (Fig. 4e, f). Furthermore, co-silencing SCOS1 and Eomes in Raptor KO γδ T cells significantly reduced of IFN-γ production, compared with silencing SOCS1 (Fig. 4g, h). Together, these results suggest that mTORC1 potentiated γδ T1 differentiation by suppressing SOCS1, which in turn negatively regulated Eomes expression.
a RNAseq heat map of differentially expressed genes in samples of WT (n = 6), Raptor KO (n = 8 mice per sample) and Rictor KO (n = 5 mice per sample) γδ T cells isolated from pooled spleen. Expression of signature genes for γδ T1 and γδ T17 were shown. b SOCS1 mRNA were analyzed by real-time quantitative PCR in WT, Raptor KO and Rictor KO γδ T cells left unstimulated or priming under γδ T1 condition for 72 h. c, d Under the γδ T1 condition, γδ T cells were transfected with SOCS1 siRNA and cells were analyzed at day 4 for IFN-γ staining. Statistics of 3–6 replicates was shown in (c). Representative FACS plots were shown in (d). e, f Eomes was stained for γδ T cells at day 4 of γδ T1 condition and the MFI of Eomes for 3–6 replicates and statistics was shown in (e) and representative Eomes staining histograms was shown in (f). g, h Under the γδ T1 condition, Raptor KO γδ T cells were transfected with SOCS1 and Eomes siRNA and cells were analyzed at day 4 for IFN-γ staining. Statistics of four replicates was shown in (g). Representative FACS plots were shown in (h). Data are means ± SEM. ns, not significant; ***P < 0.001 (two-tailed unpaired t test). Data are representative of at least three independent experiments. Numbers indicate percentage of cells in gates.
mTORC1-mediated glycolysis is required for NKG2D expression on γδ T cells
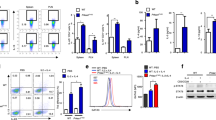
It has been reported that NKG2D expression on γδ T cells is critical for tumor immunity [27, 28]. We also analyzed NKG2D expression in γδ T cells isolated from Raptor and Rictor KO mice. The results indicated that NKG2D expression was reduced in γδ T cells from Raptor KO (Fig. 5a, b) but not Rictor KO mice (Fig. 5c, d). We then showed that the mTORC1 deficiency-mediated NKG2D reduction in γδ T cells was cell intrinsic (Fig. 5e, f). We also found that the NKG2D reduction in Raptor KO γδ T cells was independent of T-bet or Eomes (Fig. S6E–H). Because our previous study showed that rapamycin treatment in late phase of cultured γδ T cells increased NKG2D expression [19], we hypothesized that the inhibition time was responsible for this different result. To clarify this discrepancy, we added rapamycin in the early phase of γδ T cell culture. We found that mTORC1 upregulated NKG2D expression in γδ T cells at the early time points, as indicated by the suppression of NKG2D by early treatment with rapamycin (Fig. S7A, S7B). Because mTORC1 has been shown to promote glycolysis [29], we then hypothesized that glycolysis was involved in the regulation of NKG2D expression by mTORC1. We confirmed that mTORC1 was a positive regulator of glycolysis in γδ T cells (Fig. 5g). We then applied the HK2 inhibitor 2-DG (2-deoxyglucose) to inhibit glycolysis following activation with anti-γδ TCR and anti-CD28, and we observed that the reduction in NKG2D expression induced by 2-DG only occurred in glucose-free medium supplemented with glucose (Fig. 5h, i). Notably, 2-DG didn’t change NKG2D expression while γδ T cells were provided with an alternative carbon source in the absence of glucose (Fig. 5h, i), suggesting mTORC1-mediated NKG2D expression was dependent on glycolysis of glucose.
a, b Raptor-f/f and Raptor KO mice were sacrificed and γδ T cells were analyzed for NKG2D expression in spleen ex vivo. Each symbol represents an individual mouse. Statistics of 4–5 mice was shown in (a) and representative plots were shown in (b). c, d Rictor-f/f and Rictor KO mice were sacrificed and γδ T cells were analyzed for NKG2D expression in spleen ex vivo. Each symbol represents an individual mouse. Statistics of 3–6 mice was shown in (c) and representative plots were shown in (d). e, f γδ T cell NKG2D expression was analyzed for mice in Fig. 1 (p–r). Each symbol represents an individual mouse. Statistics of five mice for each group was shown in (e) and representative plots were shown in (f). g WT, Raptor KO and Rictor KO γδ T cells were expanded for 6 days and analyzed by seahorse metabolism analyzer. Statistics of triplicates of ECAR was shown in (g). h, i γδ T cells isolated from WT mice and stimulated with anti-γδ TCR and anti-CD28 condition with or without 2-DG (1 mM) in the indicated glucose (10 mM) or not or galactose (10 mM) for the first 96 h. Cells were collected and NKG2D was stained and representative plots were shown. Data are means ± SEM. ns, not significant; ***P < 0.001 (two-tailed unpaired t test). Data are representative of at least three independent experiments. Numbers indicate percentage of cells in gates.
mTORC1, but not mTORC2, is required for the cytotoxicity of γδ T cells and tumor immunity
We have previously demonstrated that γδ T cells play an important role in tumor immunity through IFN-γ and NKG2D [5, 28]. Based on the results that Raptor deficiency decreased peripheral γδ T cell IFN-γ production and NKG2D expression (Figs. 2, 5), we tested the anti-tumor function of Raptor KO γδ T cells as previously described [28]. The results indicated that Raptor KO γδ T cells had a significantly reduced killing capacity in vitro (Fig. 6a, b) and in vivo (Fig. 6c, d). We did not observe significant changes in the anti-tumor function for Rictor KO γδ T cells (Fig. S8), consistent with the results that Rictor deficiency does not affect IFN-γ or NKG2D. Therefore, our results strongly suggest that functional mTORC1 is required for γδ T cell IFN-γ production and NKG2D expression, as well as for protective anti-tumor immunity.
a γδ T cells isolated from Raptor-f/f or Raptor KO mice and expanded for 6 days (Effector cells). B16F0 target cells were labeled CFSEhi as target cells and same number of WT splenocytes were labeled CFSElo as internal cell number control. Different numbers of effector γδ T cells were mixed with the target cells. Cells were co-cultured for 12 h and remaining target cells were analyzed by FACS. Statistics of B16F0 cells percentages for triplicates was shown in (a) and representative plots were shown in (b). c, d B16F0 cells (3 × 105 cells) and expanded Raptor-f/f or Raptor KO γδ T cells (3 × 105 cells) were co-injected into flanks of TCR δ−/− mice. Raptor-f/f γδ T cells + B16F0 were injected into the left flank, and Raptor KO γδ T cells + B16F0 cells were injected into the right flank. Tumor growth for seven mice for each group was calculated and shown in (c). d Tumors for each group were isolated at day 18 post tumor injection were shown. Data are means ± SEM. ns, not significant; *P < 0.05, **P < 0.01 and ***P < 0.001(two-tailed unpaired t test). Data are representative of at least two independent experiments.
Both mTORC1 and mTORC2 are required for γδ T17 differentiation through different mechanisms
RORγt is the key transcription factor for IL-17 [11]. Consistent with Fig. 2, RORγt was markedly reduced in both Raptor KO and Rictor KO γδ T cells (Fig. 7a–d). Based on the finding that mTORC1 is a central regulator of glycolysis [29], we determined whether glycolysis was required for mTORC1-mediated γδ T17 differentiation. We found a significant reduction in glycolysis in Raptor KO γδ T17 cells (Fig. 7e). To determine if glycolysis played a role in the regulation of γδ T17, we added 2-DG to the γδ T17 culture. The results showed that the reduction of γδ T17 by 2-DG only occurred in glucose-free medium supplemented with glucose (Fig. 7f, g). Consistent with NKG2D regulation (Fig. 5h, i), 2-DG didn’t change IL-17 expression while providing with an alternative carbon source in the absence of glucose (Fig. 7f, g). Our results thus indicated that mTORC1 promotes IL-17 expression in γδ T cells though mediating glucose-dependent glycolysis.
a, b Raptor-f/f and Raptor KO mice were analyzed for γδ T cell RORγt / T-bet expression in spleen ex vivo. Each symbol represents an individual mouse. Statistics of 4–5 mice for each group was shown in (a) and representative plots were shown in (b). c, d γδ T cell RORγt and T-bet was stained for Rictor-f/f and Rictor KO mice in spleen ex vivo. Statistics of 3–5 mice for each group shown in (c) and representative plots were shown in (d). e Raptor-f/f or Raptor KO mice γδ T cells were isolated and cultured under γδ T17-skewing conditions for 5 days. Differentiated γδ T cells were analyzed by seahorse metabolism analyzer. Statistics of triplicates of ECAR was shown in (e). f, g Sorted WT γδ T cells were primed under γδ T17 condition with or without 2-deoxyglucose (2-DG) (1 mM) in the indicated glucose (10 mM) or not or galactose (10 mM) for 4 days. Cells were analyzed for IL-17 expression. h Rictor-f/f or Rictor KO γδ T cells were isolated and cultured under γδ T17 skewing conditions for 5 days. ECAR was analyzed by seahorse metabolisms analyzer. Statistics of triplicates was shown in (h). i, j Rictor-f/f or Rictor KO γδ T cells were incubated with 2′, 7′-Dichlorodihydrofluorescein diacetate (DCFH-DA) (5 μM) for 15 min at room temperature in the dark and then fluorescent 2′, 7′-dichlorofluorescein (DCF) was analyzed by FACS. Statistics of triplicates was shown in (i) and representative plots with MFI of DCF in each gate was shown in (j). k, l Rictor-f/f or Rictor KO γδ T cells were incubated with MitoSOX (5 μM) for 10 min at 37 °C in the dark and analyzed by FACS. Statistics of four duplicates were shown in (k) and representative plots with MFI of MitoSOX in each gate were shown in (l). m, n WT γδ T cells were sorted and primed under γδ T17 condition and dichloroacetate (DCA) (10 mM) was added to the culture. Cells were analyzed for IL-17 expression at day 4, with statistics of triplicates shown in (m) and representative plots were shown in (n). o, p γδ T cells from Rictor-f/f and Rictor KO mice were sorted and primed under γδ 17 condition and N-acetyl-L-cysteine (NAC) (1 mM) was added to the culture. At day 4, cells were collected and stained for IL-17. Statistics of triplicates were shown in (o) and representative plots were shown in (p). q, r γδ T cells from Rictor-f/f and Rictor KO mice were sorted and primed under γδ T17 condition and mito-tempo (Mito) (0.1 μM) was added to the culture. At day 4, cells were collected and stained for IL-17. Statistics of four duplicates were shown in (q) and representative plots were shown in (r). Data are means ± SEM. ns, not significant; *P < 0.05, **P < 0.01 and ***P < 0.001 (two-tailed unpaired t test). Numbers indicate percentage of cells in gates or quadrants.
To define the mechanism by which mTORC2 mediated γδ T17 differentiation, we first analyzed glycolysis in Rictor KO γδ T cells as described above. Interestingly, no significant change in the ECAR was observed in Rictor KO γδ T cells (Fig. 7h). Because reactive oxygen species (ROS) are known to be a negative regulator of Th17 cell differentiation [30], it is unclear whether ROS play any roles in γδ T17 differentiation. Indeed, we detected increased ROS activity in Rictor KO γδ T cells using DCF staining (Fig. 7i, j). Moreover, we found that mitoROS were also significantly enriched in mTORC2 KO γδ T cells (Fig. 7k, l). Furthermore, forced production of ROS in WT γδ T cells by treatment with dichloroacetate (DCA) decreased γδ T17 differentiation (Fig. 7m, n). To test the possibility that increased ROS activity was responsible for the reduction in IL-17 in Rictor KO γδ T cells, γδ T cells were sorted from Rictor KO mice and cultured under γδ T17 conditions in the absence or presence of ROS inhibitor, N-acetyl-L-cysteine (NAC). We found that the addition of NAC restored γδ T17 differentiation in γδ T cells from Rictor KO mice (Fig. 7o, p). Moreover, adding mito-tempo, a mitochondria-targeting superoxide dismutase mimetic with superoxide and alkyl radical scavenging properties, also restored γδ T17 differentiation to WT levels (Fig. 7q, r). Together, these results suggested that mTORC2 potentiated γδ T17 differentiation through inhibiting the production of mitoROS.
Because we found that mTORC1 and mTORC2 are both required for γδ T17 differentiation through different mechanisms, we then determined whether the two mTOR signaling pathways have potential crosstalk. By analyzing Raptor, Rictor and double KO mice ex vivo and in vitro γδ T cell differentiation assay, we found mTORC1 and mTORC2 signals synergistically promote the secretion of IL-17 in γδ T cells through different signaling pathways (Fig. S9).
Because both the thymic and extrathymic differentiation of effector γδ T cells have been reported [10, 11, 15, 16], it is not clear whether mTOR complexes play a role in thymic γδ T cell determination. Thymus analysis showed that thymic γδ T1 and γδ T17 were unchanged in Raptor KO mice. However, thymic γδ T17 was impaired in Rictor KO mice (Fig. S10). Combined with the peripheral data shown in Fig. 2a–d, these results suggested that mTORC1 is important for the peripheral differentiation and maintenance of γδ T1 and γδ T17 but that mTORC2 mainly supports γδ T17 development in the thymus.
Both mTORC1 and mTORC2 signals in γδ T cells are pathogenic in a psoriasis-like mouse model
Psoriasis is γδ T17-driven chronic skin inflammatory disease [31, 32]. Since γδ T cells from both Raptor KO and Rictor KO mice had impaired γδ T17 differentiation, we then determined whether these mice would be resistant to psoriasis. Indeed, we observed markedly reduced skin pathology (Fig. 8a–d) in both KO strains compared with the results in WT mice. We further determined that γδ T cells were the major source of IL-17 (Fig. 8e, f) and that the production of IL-17 by Raptor KO and Rictor KO γδ T cells was markedly reduced in dermal pathogenic sites (Fig. 8g, h). Moreover, we found that dermal-localization (Fig. S11A–D) and inflammation-driven recruitment (Fig. S11E–H) were both impaired in γδ T cells, especially the Vγ4 subset, from Raptor and Rictor KO mice. Collectively, our results suggested that both mTORC1 and mTORC2 participated in the pathogenesis of psoriasis through regulating γδ T cells recruitment and IL-17 production.
a, b WT, Raptor KO, and Rictor KO mice were induced psoriasis with IMQ method. At day 7 post induction, mice were sacrificed with pictures shown. c PASI score was analyzed for 5–6 mice in each group. d H&E staining for the inflamed skin was shown. e, f WT, Raptor KO and Rictor KO mice were sacrificed and dermal CD45+ cells were analyzed for γδ TCR and IL-17 staining. Each symbol represents an individual mouse. Statistics of 5 mice for each group was shown in (e). Representative plots were shown in (f). g, h Dermal γδ T cells were gated and analyzed for IFN-γ and IL-17 expression. Each symbol represents an individual mouse. Statistics of 5 mice for each group was shown in (g) and representative plots were shown in (h). Data are means ± SEM. ns, not significant; *P < 0.05, **P < 0.01 and ***P < 0.001 (two-tailed unpaired t test). Data are representative of two independent experiments. Numbers indicate percentage of cells in quadrants.
Discussion
γδ T cells have many unique features and biological functions. γδ T cells provide a cytokine milieu to regulate immunity. Therefore, understanding the molecular mechanisms that control γδ T cell cytokine production is important. In this study, we identified the function of mTOR pathways in controlling γδ Τ1 and γδ T17 differentiation as well as in regulating the effector functions of γδ T cells in tumor immunity and autoimmunity.
One of the important findings in our study was that mTORC1 was essential for IFN-γ production by γδ T cells but that mTORC2 was not necessary (Fig. 2). Using mTOR complex conditional KO mice, we demonstrated that depletion of mTORC1 impaired peripheral γδ T1 differentiation though the SOCS1-Eomes axis, which was strikingly different from that in CD4 T cells. mTORC1 suppressed SOCS1 in γδ T cells, which suppressed Eomes-mediated IFN-γ production (Fig. 4). Because the cytokine production ability of γδ T cells is partially programmed in the thymus, further studies are needed to clarify the role of Eomes in the programming of γδ T cells in the thymus.
Another notable finding of our current study was that both mTORC1 and mTORC2 were involved in γδ T17 differentiation, but through completely different mechanisms. However, the exact metabolic products of glycolysis in γδ T cells and how they trigger IL-17 differentiation and the details of how mTORC2 reduces mitoROS require further clarification.
The physiological importance of defining the roles of mTOR complexes in γδ T cell cytokine production is not fully clear. mTORC1-deficient γδ T cells lose the ability for γδ T1 differentiation and NKG2D expression (Figs. 2, 5), and consequentially, have impaired cytotoxicity against tumors both in vitro and in vivo (Fig. 6). It should be noted that although our current results seem to conflict with those of our previous report in which cultured γδ T cells with the addition of rapamycin, a specific blocking reagent of mTORC1, exhibited increases in NKG2D, TNF-α, and tumor cytotoxicity [19], we showed that the discrepancy might be due to timing of the inhibition of mTORC1 activity (Fig. S7). Our results might also help explain why the results of the usage of rapamycin or rapamycin analogs in anticancer therapy are not as good as expected, most likely because of the impairment of T cell function.
γδ T cells, especially γδ T17 cells, have also been implicated in inflammatory pathology. Interestingly, IMQ-induced psoriasis-like skin inflammation was significantly impaired in both Raptor and Rictor KO mice (Fig. 8), most likely because of the impairment of IL-17 production by γδ T cells (Fig. 8). Further studies are needed to determine whether γδ T cell functions are altered in patients with psoriasis, and the manipulation of γδ T cells may lead to novel therapies for psoriasis.
In summary, we identified previously unknown functions of mTORC1 and mTORC2 in γδ T cell differentiation and clarified their different roles in mediating the function of γδ T cells against tumors and in autoimmunity. Our work will help guide future efforts to manipulate γδ T cells by altering mTORC1 or mTORC2 activity for use in immunotherapy.
Materials and methods
Mice
B6.Cg-Rptortm1.1Dmsa/J (B6 Raptor-flox), Rictortm1.1Klg/SjmJ (B6 Rictor-flox), B6.Cg-Tg(CD2-icre)4Kio/J (hCD2-iCre) and B6.129P2-Tcrdtm1Mom/J (TCR δ−/−) mice are purchased from The Jackson Laboratory (Bar Harbor, ME). Eomes-flox, T-bet-flox and CD45.1 mice were given by Zhongjun Dong from Tsinghua University (Beijing, China). Sex- and Age-matched animals were randomly assigned to different groups. All mice were kept in SPF condition and mice protocols were approved by the animal experiment committee of Jinan University.
Reagents
Purified anti-mouse TCR γδ mAb (UC7), anti-mouse CD3 mAb (145-2C11), anti-mouse CD28 mAb (PV1), purified anti-mouse IFN-γ (XMG1.2), PE-conjugated anti-mouse Vγ1 mAb (2.11), APC/PE-conjugated anti-mouse IL-17 mAb (17F3), APC-conjugated anti-mouse CD8 mAb (53.6.7) and APC-conjugated anti-mouse T-bet (4B10) were purchased from Sungen (Tianjin, China). APC/Percpcy5.5-conjugated anti-mouse CD4 (GK1.5), FITC/APC-conjugated anti-mouse TCR γδ mAb (GL3), PE-Cy7-conjugated anti-mouse IFN-γ mAb (XMG1.2), PE-Cy7-conjugated anti-mouse CD3 mAb (145-2C11), APC-conjugated anti-mouse Vγ4 mAb (UC3-10A6) and PE-conjugated anti-mouse Ki67(16A8) were purchased from Biolegend (San Diego, CA, USA). PE-conjugated anti-mouse CD98 (RL388), APC-conjugated anti-mouse CD71 (R17217), eFlour450-conjugated anti-mouse CD3 (17A2), PE-conjugated anti-mouse NKG2D (CX5), PE-conjugated anti-mouse Eomes (Dan11mag), APC-conjugated anti-mouse RORγt (B2D), Percpcy5.5-conjugated anti-mouse IL-17 mAb (eBio17B7) and Foxp3 staining buffer set were purchased from eBioscience (San Diego, CA, USA). PE-conjugated anti-mouse pAkt473 mAb (M89-61) was purchased from BD Biosciences (Franklin Lakes, NJ, USA). Purified anti-mouse Raptor mAb (24C12), purified anti-mouse Rictor mAb (53A2), 488-conjugated anti-human/mouse pS6 mAb (D57.2.2E), PE-conjugated anti-mouse cleaved casp-3 mAb (5A1E) and 647-conjugated anti-mouse Bim mAb (C34C5) were purchased from Cell Signaling Technology (Danvers, MA, USA).
Tumor models
B16F0 melanoma cells were mixed with γδ T cells and injected s.c., and tumor growth was monitored and recorded daily as described previously [28, 33]. Sample size was chosen to ensure the possibility of statistical analysis and to also minimize the use of animals in accordance with animal experiment committee of Jinan University. The results from previous results were also used to determine the sample size.
Imiquimod (IMQ)-induced psoriasis
Female Raptor-f/f, Rictor-f/f, Raptor KO, and Rictor KO mice (7 weeks of age) were maintained under SPF conditions. The mice were treated with a daily topical dose of 62.5 mg of IMQ cream (Sichuan Med-shine Pharmaceutical, H20030128) on their shaved backs for 7 consecutive days. After the mice were sacrificed, part of the skin sample was fixed in 4% paraformaldehyde for H&E staining, and the rest was used to isolate dermal lymphocytes for intracellular cytokine staining. Sample size was chosen to ensure the possibility of statistical analysis and to also minimize the use of animals in accordance with animal experiment committee of Jinan University. The results from previous results were also used to determine the sample size.
Cell preparation, activation, and staining
γδ T cells were sorted and expanded as previously described [28, 33]. For the expansion of γδ T cells, day 6-γδ T cells were used, and the purity was checked (>99%). For in vitro differentiation, sorted γδ T cells were cultured in anti-γδ TCR (clone UC7)-coated plates (10 μg/ml) with soluble anti-CD28 (clone 37.51) (1 μg/ml) and IL-2 (2 ng/ml). IL-12 (5 ng/ml) and anti-IL-4 (clone 11B11) (10 μg/ml) were added for γδ T1-skewing conditions, and TGF-β (2 ng/ml), IL-6 (5 ng/ml), and anti-IFN-γ (clone XMG1.2) (5 μg/ml) were added for γδ T17-skewing conditions. For intracellular cytokine staining, the protocols described in our previous publications were used [28, 33]. FACS was performed with BD FACSVerse, and data were analyzed with Flowjo 7.0 software.
In vitro killing assay
B16F0 cells (8000 cells/well) were labeled with CFSE (5 μM). Splenocytes were labeled with CFSE (0.25 μM) as an internal cell number control. Then, B16F0 cells and splenocytes were mixed with expanded γδ T cells for 12 h. Killing capability was assessed by determining the percentage of remaining B16F0 cells (% of CFSEhi cells).
Real-time PCR for gene transcription
Total RNA was extracted from cells and reverse-transcribed with a Takara reverse transcription kit. The primers for the mouse genes were: mTOR-F: CAGTTCGCCAGTGGACTGAAG, mTOR-R: GCTGGTCATAGAAGCGAGTAGAC, Raptor-F: TTTGTCTACGACTGTTCCAATGC, Raptor-R: GCTACCTCTAGTTCCTGCTCC, Rictor-F: ACAGTTGGAAAAGTGGCACAA, Rictor-R: GCGACGAACGTAGTTATCACCA, SOCS1-F: CTGCGGCTTCTATTGGGGAC, SOCS1-R: AAAAGGCAGTCGAAGGTCTCG. The primers for internal control gene β-Actin were Actb-F: AACAGTCCGCCTAGAAGCAC, Actb-R: CGTTGACATCCGTAAAGACC. Real-time PCR was performed with a BioRad CFX Connect cycler.
RNA sequencing
The whole cDNA products were generated using the Discover-scTM WTA Kit V2 (Vazyme, N711). The sequencing libraries were generated using the TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme, TD503). Library preparations were sequenced on the Illumina Hiseq X Ten platform with the 150 bp paired-end module. The RNAseq data were deposited in the Sequence Read Archive (SRA) repository at NCBI under the accession number SRP214746.
RNA interference
RNAi was performed as described previously [34]. Sequences for the siRNAs were: The siControl sense: UUCUCCGAACGUGUCACGUTT, siControl anti-sense: ACGUGACACGUUCGGAGAATT. siSOCS1 includes three oligonucleotides pairs: sense: ACACUCACUUCCGCACCUUTT, anti-sense: AAGGUGCGGAAGUGAGUGUTT, sense: CUCAGCGUGAAGAUGGCUUTT, anti-sense: AAGCCAUCUUCACGCUGAGTT, sense: CUCCGUGACUACCUGAGUUTT, anti-sense: AACUCAGGUAGUCACGGAGTT. siEomes includes two oligonucleotides pairs: sense: GGCUCUUAUUUCUACUCAU, anti-sense: CCGAGAAUAAAGAUGAGUA, sense: GCAAUAAGAUGUACGUUCA, anti-sense: CGUUAUUCUACAUGCAAGU.
Statistics
Statistical significance was calculated by two-tailed unpaired Student t test using GraphPad Prism 6 for Windows (GraphPad). The following terminology is used to show statistical significance: *p < 0.05, **p < 0.01, ***p < 0.001. No samples or animals were excluded from the analysis. Variances are similar between the groups in t test analysis.
References
Born WK, Yin Z, Hahn YS, Sun D, O’Brien RL. Analysis of gamma delta T cell functions in the mouse. J Immunol. 2010;184:4055–61.
Papotto PH, Ribot JC, Silva-Santos B. IL-17(+) gammadelta T cells as kick-starters of inflammation. Nat Immunol. 2017;18:604–11.
Silva-Santos B, Serre K, Norell H. gammadelta T cells in cancer. Nat Rev Immunol. 2015;15:683–91.
Carding SR, Egan PJ. Gammadelta T cells: functional plasticity and heterogeneity. Nat Rev Immunol. 2002;2:336–45.
Gao Y, Yang W, Pan M, Scully E, Girardi M, Augenlicht LH, et al. Gamma delta T cells provide an early source of interferon gamma in tumor immunity. J Exp Med. 2003;198:433–42.
Hiromatsu K, Yoshikai Y, Matsuzaki G, Ohga S, Muramori K, Matsumoto K, et al. A protective role of gamma/delta T cells in primary infection with Listeria monocytogenes in mice. J Exp Med. 1992;175:49–56.
Yin Z, Zhang DH, Welte T, Bahtiyar G, Jung S, Liu L, et al. Dominance of IL-12 over IL-4 in gamma delta T cell differentiation leads to default production of IFN-gamma: failure to down-regulate IL-12 receptor beta 2-chain expression. J Immunol. 2000;164:3056–64.
Yin Z, Chen C, Szabo SJ, Glimcher LH, Ray A, Craft J. T-Bet expression and failure of GATA-3 cross-regulation lead to default production of IFN-gamma by gammadelta T cells. J Immunol. 2002;168:1566–71.
Chen L, He W, Kim ST, Tao J, Gao Y, Chi H, et al. Epigenetic and transcriptional programs lead to default IFN-gamma production by gammadelta T cells. J Immunol. 2007;178:2730–6.
Jensen KD, Su X, Shin S, Li L, Youssef S, Yamasaki S, et al. Thymic selection determines gammadelta T cell effector fate: antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon gamma. Immunity. 2008;29:90–100.
Ribot JC, deBarros A, Pang DJ, Neves JF, Peperzak V, Roberts SJ, et al. CD27 is a thymic determinant of the balance between interferon-gamma- and interleukin 17-producing gammadelta T cell subsets. Nat Immunol. 2009;10:427–36.
Munoz-Ruiz M, Ribot JC, Grosso AR, Goncalves-Sousa N, Pamplona A, Pennington DJ, et al. TCR signal strength controls thymic differentiation of discrete proinflammatory gammadelta T cell subsets. Nat Immunol. 2016;17:721–7.
Sumaria N, Grandjean CL, Silva-Santos B, Pennington DJ. Strong TCRgammadelta signaling prohibits thymic development of IL-17A-secreting gammadelta T cells. Cell Rep. 2017;19:2469–76.
Spidale NA, Sylvia K, Narayan K, Miu B, Frascoli M, Melichar HJ, et al. Interleukin-17-Producing gammadelta T Cells Originate from SOX13(+) Progenitors that Are Independent of gammadeltaTCR Signaling. Immunity. 2018;49:857–872.e855.
Papotto PH, Goncalves-Sousa N, Schmolka N, Iseppon A, Mensurado S, Stockinger B, et al. IL-23 drives differentiation of peripheral gammadelta17 T cells from adult bone marrow-derived precursors. EMBO Rep. 2017;18:1957–67.
Muschaweckh A, Petermann F, Korn T. IL-1beta and IL-23 promote extrathymic commitment of CD27(+)CD122(−) gammadelta T cells to gammadeltaT17 cells. J Immunol. 2017;199:2668–79.
Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–38.
Yang K, Blanco DB, Chen X, Dash P, Neale G, Rosencrance C, et al. Metabolic signaling directs the reciprocal lineage decisions of alphabeta and gammadelta T cells. Sci Immunol. 2018;3. pii: eaas9818. https://doi.org/10.1126/sciimmunol.aas9818.
Cao G, Wang Q, Li G, Meng Z, Liu H, Tong J, et al. mTOR inhibition potentiates cytotoxicity of Vgamma4 gammadelta T cells via up-regulating NKG2D and TNF-alpha. J Leukoc Biol. 2016;100:1181–9.
Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu Rev Immunol. 2012;30:39–68.
Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol Rev. 2012;249:43–58.
Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303.
Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, et al. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–53.
Lazarevic V, Glimcher LH, Lord GM. T-bet: a bridge between innate and adaptive immunity. Nat Rev Immunol. 2013;13:777–89.
Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–44.
Barros-Martins J, Schmolka N, Fontinha D, Pires de Miranda M, Simas JP, Brok I, et al. Effector gammadelta T cell differentiation relies on master but not auxiliary Th cell transcription factors. J Immunol. 2016;196:3642–52.
Kabelitz D, Wesch D, He W. Perspectives of gammadelta T cells in tumor immunology. Cancer Res. 2007;67:5–8.
He W, Hao J, Dong S, Gao Y, Tao J, Chi H, et al. Naturally activated V gamma 4 gamma delta T cells play a protective role in tumor immunity through expression of eomesodermin. J Immunol. 2010;185:126–33.
Altomare DA, Khaled AR. Homeostasis and the importance for a balance between AKT/mTOR activity and intracellular signaling. Curr Med Chem. 2012;19:3748–62.
Fu G, Xu Q, Qiu Y, Jin X, Xu T, Dong S, et al. Suppression of Th17 cell differentiation by misshapen/NIK-related kinase MINK1. J Exp Med. 2017;214:1453–69.
Brembilla NC, Senra L, Boehncke WH. The IL-17 family of cytokines in psoriasis: IL-17A and Beyond. Front Immunol. 2018;9:1682.
Cai Y, Shen X, Ding C, Qi C, Li K, Li X, et al. Pivotal role of dermal IL-17-producing gammadelta T cells in skin inflammation. Immunity. 2011;35:596–610.
Hao J, Dong S, Xia S, He W, Jia H, Zhang S, et al. Regulatory role of Vgamma1 gammadelta T cells in tumor immunity through IL-4 production. J Immunol. 2011;187:4979–86.
Zhu E, Wang X, Zheng B, Wang Q, Hao J, Chen S, et al. miR-20b suppresses Th17 differentiation and the pathogenesis of experimental autoimmune encephalomyelitis by targeting RORgammat and STAT3. J Immunol. 2014;192:5599–609.
Acknowledgements
The authors thank Professor Zhongjun Dong from Tsinghua University (Beijing, China) for giving us T-bet-flox and Eomes-flox mice. We thank Qiong Wen for managing the mice care in our facility. We thank Bean Liu from Jinan University (Guangzhou, China) for editing the language of the paper. This work was supported by the Grants from National Natural Science Foundation of China (31420103901 to Z.Y., 81630025 to J.H., 31700753 to G.C., and 31700768 to Q.L.), Grant from Traditional Chinese Medicine Bureau of Guangdong Province (2018071 to J.H.), and Grants from China Postdoctoral Science Foundation (2017M612840 to G.C., 2017M612841 to Q.L., and 2018M643375 to J.L.), Grant from Guangzhou Municipal Science and Technology Bureau (201904010090 to J.H.) and Grant from Health Commission of Guangdong Province (A2019520 to J.H.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by G. Del Sal
Rights and permissions
About this article
Cite this article
Yang, Q., Liu, X., Liu, Q. et al. Roles of mTORC1 and mTORC2 in controlling γδ T1 and γδ T17 differentiation and function. Cell Death Differ 27, 2248–2262 (2020). https://doi.org/10.1038/s41418-020-0500-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-020-0500-9
This article is cited by
-
Landscape of unconventional γδ T cell subsets in cancer
Molecular Biology Reports (2024)
-
Gamma delta T-cell-based immune checkpoint therapy: attractive candidate for antitumor treatment
Molecular Cancer (2023)
-
γδ T cells: origin and fate, subsets, diseases and immunotherapy
Signal Transduction and Targeted Therapy (2023)
-
Artesunate targets cellular metabolism to regulate the Th17/Treg cell balance
Inflammation Research (2023)
-
Complex Interplay Between Metabolism and CD4+ T-Cell Activation, Differentiation, and Function: a Novel Perspective for Atherosclerosis Immunotherapy
Cardiovascular Drugs and Therapy (2023)