Abstract
Main conclusion
The structure of the cotton uceA1.7 promoter and its modules was analyzed; the potential of their key sequences has been confirmed in different tissues, proving to be a good candidate for the development of new biotechnological tools.
Abstract
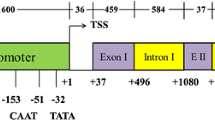
Transcriptional promoters are among the primary genetic engineering elements used to control genes of interest (GOIs) associated with agronomic traits. Cotton uceA1.7 was previously characterized as a constitutive promoter with activity higher than that of the constitutive promoter from the Cauliflower mosaic virus (CaMV) 35S gene in various plant tissues. In this study, we generated Arabidopsis thaliana homozygous events stably overexpressing the gfp reporter gene driven by different modules of the uceA1.7 promoter. The expression level of the reporter gene in different plant tissues and the transcriptional stability of these modules was determined compared to its full-length promoter and the 35S promoter. The full-length uceA1.7 promoter exhibited higher activity in different plant tissues compared to the 35S promoter. Two modules of the promoter produced a low and unstable transcription level compared to the other promoters. The other two modules rich in cis-regulatory elements showed similar activity levels to full-length uceA1.7 and 35S promoters but were less stable. This result suggests the location of a minimal portion of the promoter that is required to initiate transcription properly (the core promoter). Additionally, the full-length uceA1.7 promoter containing the 5′-untranslated region (UTR) is essential for higher transcriptional stability in various plant tissues. These findings confirm the potential use of the full-length uceA1.7 promoter for the development of new biotechnological tools (NBTs) to achieve higher expression levels of GOIs in, for example, the root or flower bud for the efficient control of phytonematodes and pest-insects, respectively, in important crops.



Similar content being viewed by others
Abbreviations
- GOIs:
-
Genes of interest
- NBTs:
-
New biotechnological tools
- CaMV:
-
Cauliflower mosaic virus
- UTR:
-
Untranslated region
- TSS:
-
Transcriptional start site
References
Acharya S, Ranjan R, Pattanaik S, Maiti IB, Dey N (2014) Efficient chimeric plant promoters derived from plant infecting viral promoter sequences. Planta 239(2):381–396. https://doi.org/10.1007/s00425-013-1973-2
Artico S, Lambret-Frotté J, Nardeli SM, Oliveira-Neto OB, Grossi-de-Sa MF, Alves-Ferreira M (2014) Isolation and characterization of three new promoters from Gossypium hirsutum that show high activity in reproductive tissues. Plant Mol Biol Rep 32:630–643
Basso MF, Ferreira PCG, Kobayashi AK, Harmon FG, Nepomuceno AL, Molinari HBC, Grossi-de-Sa MF (2019) MicroRNAs and new biotechnological tools for its modulation and improving stress tolerance in plants. Plant Biotechnol J 17:1482–1500. https://doi.org/10.1111/pbi.13116
Benfey PN, Chua NH (1990) The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250(4983):959–966. https://doi.org/10.1126/science.250.4983.959
Benfey PN, Ren L, Chua NH (1990) Tissue-specific expression from CaMV 35S enhancer subdomains in early stages of plant development. EMBO J 9(6):1677–1684
Biłas R, Szafran K, Hnatuszko-Konka K, Kononowicz AK (2016) Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Cult 127(2):269–287. https://doi.org/10.1007/s11240-016-1057-7
Bulger M, Groudine M (2011) Functional and mechanistic diversity of distal transcription enhancers. Cell 144(3):327–339. https://doi.org/10.1016/j.cell.2011.01.024
Burley SK, Roeder RG (1996) Biochemistry and structural biology of transcription factor IID (TFIID). Annu Rev Biochem 65:769–799. https://doi.org/10.1146/annurev.bi.65.070196.004005
Chang WC, Lee TY, Huang HD, Huang HY, Pan RL (2008) PlantPAN: plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene groups. BMC Genom 9:561. https://doi.org/10.1186/1471-2164-9-561
Chow CN, Lee TY, Hung YC, Li GZ, Tseng KC, Liu YH, Kuo PL, Zheng HQ, Chang WC (2019) PlantPAN3.0: a new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res 47(D1):D1155–D1163. https://doi.org/10.1093/nar/gky1081
Christensen AH, Sharrock RA, Quail PH (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 18(4):675–689
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743
Conci LR, Nishizawa Y, Saito M, Date T, Hasegawa A, Miki K, Hibi T (1993) A strong promoter fragment from the large noncoding region of soybean chlorotic mottle virus DNA. Jpn J Phytopath 59(4):432–437. https://doi.org/10.3186/jjphytopath.59.432
Crisp PA, Ganguly D, Eichten SR, Borevitz JO, Pogson BJ (2016) Reconsidering plant memory: intersections between stress recovery, RNA turnover, and epigenetics. Sci Adv 2(2):e1501340. https://doi.org/10.1126/sciadv.1501340
Diamos AG, Mason HS (2018) Chimeric 3’ flanking regions strongly enhance gene expression in plants. Plant Biotechnol J 16(12):1971–1982. https://doi.org/10.1111/pbi.12931
Dong OX, Ronald PC (2019) Genetic engineering for disease resistance in plants: recent progress and future perspectives. Plant Physiol 180(1):26–38. https://doi.org/10.1104/pp.18.01224
Fukuoka H, Ogawa T, Mitsuhara I, Iwai T, Isuzugawa K, Nishizawa Y, Gotoh Y, Nishizawa Y, Tagiri A, Ugaki M, Ohshima M, Yano H, Murai N, Niwa Y, Hibi T, Ohashi Y (2000) Agrobacterium-mediated transformation of monocot and dicot plants using the NCR promoter derived from soybean chlorotic mottle virus. Plant Cell Rep 19(8):815–820. https://doi.org/10.1007/s002990000191
Garbarino JE, Oosumi T, Belknap WR (1995) Isolation of a polyubiquitin promoter and its expression in transgenic potato plants. Plant Physiol 109(4):1371–1378. https://doi.org/10.1104/pp.109.4.1371
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40((Database issue)):D1178–D1186. https://doi.org/10.1093/nar/gkr944
Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64(2):355–365. https://doi.org/10.1111/j.1365-313X.2010.04322.x
Gupta P, Raghuvanshi S, Tyagi AK (2001) Assessment of the efficiency of various gene promoters via biolistics in leaf and regenerating seed callus of millets, Eleusine coracana and Echinochloa crusgalli. Plant Biotechnol J 18(4):275–282. https://doi.org/10.5511/plantbiotechnology.18.275
Harper G, Hull R (1998) Cloning and sequence analysis of banana streak virus DNA. Virus Genes 17(3):271–278. https://doi.org/10.1023/a:1008021921849
Harrison SJ, Mott EK, Parsley K, Aspinall S, Gray JC, Cottage A (2006) A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2(1):19. https://doi.org/10.1186/1746-4811-2-19
Hasegawa A, Verver J, Shimada A, Saito M, Goldbach R, Van Kammen A, Miki K, Kameya-Iwaki M, Hibi T (1989) The complete sequence of soybean chlorotic mottle virus DNA and the identification of a novel promoter. Nucleic Acids Res 17(23):9993–10013
Hernandez-Garcia CM, Finer JJ (2014) Identification and validation of promoters and cis-acting regulatory elements. Plant Sci 217–218:109–119. https://doi.org/10.1016/j.plantsci.2013.12.007
Hernandez-Garcia CM, Martinelli AP, Bouchard RA, Finer JJ (2009) A soybean (Glycine max) polyubiquitin promoter gives strong constitutive expression in transgenic soybean. Plant Cell Rep 28(5):837–849. https://doi.org/10.1007/s00299-009-0681-7
Hickey LT, Hafeez AN, Robinson H, Jackson SA, Leal-Bertioli SCM, Tester M, Gao C, Godwin ID, Hayes BJ, Wulff BBH (2019) Breeding crops to feed 10 billion. Nat Biotechnol 37(7):744–754. https://doi.org/10.1038/s41587-019-0152-9
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27(1):297–300. https://doi.org/10.1093/nar/27.1.297
Lambret-Frotte J, Artico S, Muniz Nardeli S, Fonseca F, Brilhante Oliveira-Neto O, Grossi-de-Sa MF, Alves-Ferreira M (2016) Promoter isolation and characterization of GhAO-like1, a Gossypium hirsutum gene similar to multicopper oxidases that is highly expressed in reproductive organs. Genome 59(1):23–36. https://doi.org/10.1139/gen-2015-0055
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327. https://doi.org/10.1093/nar/30.1.325
Lim S-H, Kim JK, Lee J-Y, Kim Y-M, Sohn S-H, Kim D-H, Ha S-H (2013) Petal-specific activity of the promoter of an anthocyanidin synthase gene of tobacco (Nicotiana tabacum L.). Plant Cell Tissue Organ Cult 114(3):373–383. https://doi.org/10.1007/s11240-013-0332-0
Limera C, Sabbadini S, Sweet JB, Mezzetti B (2017) New biotechnological tools for the genetic improvement of major woody fruit species. Front Plant Sci 8:1418. https://doi.org/10.3389/fpls.2017.01418
Liu W, Stewart CN Jr (2016) Plant synthetic promoters and transcription factors. Curr Opin Biotechnol 37:36–44. https://doi.org/10.1016/j.copbio.2015.10.001
Lu J, Sivamani E, Azhakanandam K, Samadder P, Li X, Qu R (2008a) Gene expression enhancement mediated by the 5' UTR intron of the rice rubi3 gene varied remarkably among tissues in transgenic rice plants. Mol Genet Genom 279(6):563–572. https://doi.org/10.1007/s00438-008-0333-6
Lu J, Sivamani E, Li X, Qu R (2008b) Activity of the 5’ regulatory regions of the rice polyubiquitin rubi3 gene in transgenic rice plants as analyzed by both GUS and GFP reporter genes. Plant Cell Rep 27(10):1587–1600. https://doi.org/10.1007/s00299-008-0577-y
Ma S, Shah S, Bohnert HJ, Snyder M, Dinesh-Kumar SP (2013) Incorporating motif analysis into gene co-expression networks reveals novel modular expression pattern and new signaling pathways. PLoS Genet 9(10):e1003840. https://doi.org/10.1371/journal.pgen.1003840
Maiti IB, Gowda S, Kiernan J, Ghosh SK, Shepherd RJ (1997) Promoter/leader deletion analysis and plant expression vectors with the Figwort mosaic virus (FMV) full length transcript (FLt) promoter containing single or double enhancer domains. Transgenic Res 6(2):143–156. https://doi.org/10.1023/a:1018477705019
Molina C, Grotewold E (2005) Genome wide analysis of Arabidopsis core promoters. BMC Genom 6:1–12. https://doi.org/10.1186/1471-2164-6-25
Moon J, Parry G, Estelle M (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16(12):3181–3195. https://doi.org/10.1105/tpc.104.161220
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15(3):473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Odell JT, Nagy F, Chua NH (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313(6005):810–812. https://doi.org/10.1038/313810a0
Park S-H, Yi N, Kim YS, Jeong M-H, Bang S-W, Choi YD, Kim J-K (2010) Analysis of five novel putative constitutive gene promoters in transgenic rice plants. J Exp Bot 61(9):2459–2467. https://doi.org/10.1093/jxb/erq076
Ribeiro TP, Arraes FBM, Lourenco-Tessutti IT, Silva MS, Lisei-de-Sa ME, Lucena WA, Macedo LLP, Lima JN, Santos Amorim RM, Artico S, Alves-Ferreira M, Mattar Silva MC, Grossi-de-Sa MF (2017) Transgenic cotton expressing Cry10Aa toxin confers high resistance to the cotton boll weevil. Plant Biotechnol J 15(8):997–1009. https://doi.org/10.1111/pbi.12694
Sanger M, Daubert S, Goodman RM (1990) Characteristics of a strong promoter from figwort mosaic virus: comparison with the analogous 35S promoter from cauliflower mosaic virus and the regulated mannopine synthase promoter. Plant Mol Biol 14(3):433–443. https://doi.org/10.1007/bf00028779
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101. https://doi.org/10.1038/nprot.2008.73
Smale ST, Kadonaga JT (2003) The RNA polymerase II core promoter. Annu Rev Biochem 72:449–479. https://doi.org/10.1146/annurev.biochem.72.121801.161520
Somssich M (2019) A short history of the CaMV 35S promoter. PeerJ Prepr 7:e27096v27093. https://doi.org/10.7287/peerj.preprints.27096v3
Stavolone L, Kononova M, Pauli S, Ragozzino A, de Haan P, Milligan S, Lawton K, Hohn T (2003) Cestrum yellow leaf curling virus (CmYLCV) promoter: a new strong constitutive promoter for heterologous gene expression in a wide variety of crops. Plant Mol Biol 53(5):663–673. https://doi.org/10.1023/B:PLAN.0000019110.95420.bb
Tzafrir I, Torbert KA, Lockhart BE, Somers DA, Olszewski NE (1998) The sugarcane bacilliform badnavirus promoter is active in both monocots and dicots. Plant Mol Biol Rep 38(3):347–356
van der Meer IM, Spelt CE, Mol JNM, Stuitje AR (1990) Promoter analysis of the chalcone synthase (chsA) gene of Petunia hybrida: a 67 bp promoter region directs flower-specific expression. Plant Mol Biol 15(1):95–109. https://doi.org/10.1007/bf00017727
van der Meer IM, Brouwer M, Spelt CE, Mol JN, Stuitje AR (1992) The TACPyAT repeats in the chalcone synthase promoter of Petunia hybrida act as a dominant negative cis-acting module in the control of organ-specific expression. Plant J 2(4):525–535
Viana AA, Fragoso RR, Guimaraes LM, Pontes N, Oliveira-Neto OB, Artico S, Nardeli SM, Alves-Ferreira M, Batista JA, Silva MC, Grossi-de-Sa MF (2011) Isolation and functional characterization of a cotton ubiquitination-related promoter and 5’UTR that drives high levels of expression in root and flower tissues. BMC Biotechnol 11:115. https://doi.org/10.1186/1472-6750-11-115
Wang J, Oard JH (2003) Rice ubiquitin promoters: deletion analysis and potential usefulness in plant transformation systems. Plant Cell Rep 22(2):129–134. https://doi.org/10.1007/s00299-003-0657-y
Wang R, Zhu M, Ye R, Liu Z, Zhou F, Chen H, Lin Y (2015) Novel green tissue-specific synthetic promoters and cis-regulatory elements in rice. Sci Rep 5:18256. https://doi.org/10.1038/srep18256
Wang WZ, Yang BP, Feng XY, Cao ZY, Feng CL, Wang JG, Xiong GR, Shen LB, Zeng J, Zhao TT, Zhang SZ (2017) Development and characterization of transgenic sugarcane with insect resistance and herbicide tolerance. Front Plant Sci 8:1535–1535. https://doi.org/10.3389/fpls.2017.01535
Weeks JT, Anderson OD, Blechl AE (1993) Rapid production of multiple independent lines of fertile transgenic wheat (Triticum aestivum). Plant Physiol 102(4):1077–1084. https://doi.org/10.1104/pp.102.4.1077
Wei H, Wang ML, Moore PH, Albert HH (2003) Comparative expression analysis of two sugarcane polyubiquitin promoters and flanking sequences in transgenic plants. J Plant Physiol 160(10):1241–1251. https://doi.org/10.1078/0176-1617-01086
Yamamoto YY, Ichida H, Matsui M, Obokata J, Sakurai T, Satou M, Seki M, Shinozaki K, Abe T (2007) Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genom 8:67. https://doi.org/10.1186/1471-2164-8-67
Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33(5):531–537. https://doi.org/10.1038/nbt.3207
Zhou B, Mural RV, Chen X, Oates ME, Connor RA, Martin GB, Gough J, Zeng L (2017) A subset of ubiquitin-conjugating enzymes is essential for plant immunity. Plant Physiol 173(2):1371–1390. https://doi.org/10.1104/pp.16.01190
Acknowledgements
We are grateful to EMBRAPA, CAPES, CNPq, INCT PlantStress Biotech, and FAP-DF for providing financial support for this scientific research.
Funding
MFB is grateful to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for a post-doctoral research fellowship (PDJ 150936/2018-4). This work was supported by grants from CAPES, CNPq, FAP-DF, INCT, and EMBRAPA.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2020_3348_MOESM1_ESM.tif
Supplementary file1 (TIF 6714 kb) Overview of the four uceA1.7 modules (highlighted in bold background) aligned with the full-length uceA1.7 promoter. a Module 1 containing 681 bp. b Module 2 containing 792 bp. c Module 3 containing 581 bp. d Module 4 containing 251 bp were planned around the core promoter elements predicted in this study or by Viana et al. (2011). ORF, open reading frame; SC, translation start codon; TSS, transcription start site; 5’ UTR, 5’ untranslated region
425_2020_3348_MOESM2_ESM.tif
Supplementary file2 (TIF 9023 kb) Genetic transformation of Arabidopsis thaliana mediated by Agrobacterium tumefaciens GV3101 strain. a Six independent events from each full-length promoter (uceA1.7 and CaMV 35S) or four uceA1.7 modules (Mod 1 to 4) were evaluated. b PCR detection of the transgene in A. thaliana T1 events using specific primers for the gfp (green fluorescent protein) reporter gene (Suppl. Table 1). Marker: 1.0-kb DNA ladder (Invitrogen® Cat. #10787018); WT, wild-type A. thaliana line used as a negative control for PCR and RT-qPCR assays; C+, DNA plasmid used as a positive control for the PCR assay
Rights and permissions
About this article
Cite this article
Basso, M.F., Lourenço-Tessutti, I.T., Busanello, C. et al. Insights obtained using different modules of the cotton uceA1.7 promoter. Planta 251, 56 (2020). https://doi.org/10.1007/s00425-020-03348-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-020-03348-8