Abstract
Coupling microbial electrosynthesis to renewable energy sources can provide a promising future technology for carbon dioxide conversion. However, this technology suffers from a limited number of suitable biocatalysts, resulting in a narrow product range. Here, we present the characterization of the first thermoacidophilic electroautotrophic community using chronoamperometric, metagenomic, and 13C-labeling analyses. The cathodic biofilm showed current consumption of up to −80 µA cm−2 over a period of 90 days (−350 mV vs. SHE). Metagenomic analyses identified members of the genera Moorella, Desulfofundulus, Thermodesulfitimonas, Sulfolobus, and Acidianus as potential primary producers of the biofilm, potentially thriving via an interspecies sulfur cycle. Hydrogenases seem to be key for cathodic electron uptake. An isolation campaign led to a pure culture of a Knallgas bacterium from this community. Growth of this organism on cathodes led to increasing reductive currents over time. Transcriptomic analyses revealed a distinct gene expression profile of cells grown at a cathode. Moreover, pressurizable flow cells combined with optical coherence tomography allowed an in situ observation of cathodic biofilm growth. Autotrophic growth was confirmed via isotope analysis. As a natural polyhydroxybutyrate (PHB) producer, this novel species, Kyrpidia spormannii, coupled the production of PHB to CO2 fixation on cathode surfaces.
Similar content being viewed by others
Introduction
Isolation campaigns are a valid tool to identify new biocatalysts with a specific physiological capability under conditions that typically resemble the conditions of an envisioned future biotechnological process. Moreover, isolation campaigns can also offer the chance to identify new physiological traits and interactions within a microbial community. This study describes the enrichment of an extremophilic microbial community on a cathode surface with the aim to isolate species that can thrive in the form of a biofilm using carbon dioxide as carbon source and a cathode directly or indirectly as electron and energy source.
Anthropogenic carbon dioxide emissions are stated as the main cause for climate change and a transition toward a bio-based, circular economy is widely seen as inevitable [1]. The application of CO2 as carbon source for the production of platform chemicals is a crucial step within this envisioned circular economy [1,2,3,4]. Autotrophic organisms have developed strategies for the utilization of light and chemical energy to sustain carbon dioxide fixation. In 2009, Cheng et al. provided evidence that some microorganisms could also use electrical energy as primary source of electrons and energy for carbon dioxide fixation [5]. This process, later called microbial electrosynthesis (MES) [6], has the potential to be a new sustainable form of carbon dioxide usage and energy storage, especially if the electrical energy originates from renewables [7,8,9,10].
Interestingly, the underlying processes enabling the import of electrons from cathodes are so far poorly understood and the definition of electroautotrophic growths is not clear. It is still under debate if (I) electron transfer from a cathode is mediated via cathode-driven hydrogen production, (II) occurs directly via enzymatic electron uptake and transport into the respiratory chain, or (III) is based on an enzymatic activity that catalyzes the production of an intermediary electron donor like hydrogen or formate on the electrode surface which is ultimately used by the organisms [11,12,13,14,15,16]. The standard redox potential E°′ of the hydrogen evolution reaction is −413 mV at pH 7. As a proton dependent reaction, this redox potential shifts to more positive values under acidic conditions (−206 mV at pH 3.5). Hence, acidophilic hydrogenotrophic microorganisms could have a growth benefit at a given cathode potential compared with neutrophiles. Still, the ability to grow lithoautotrophically using hydrogen as an electron donor, does not seem to be the only requirement for biofilms thriving on cathodes. Instead, the biocatalyst’s ability to cope with for instance reactive oxygen species that are produced at the electrodes seems to be one crucial factor, especially when working in oxic systems [17]. The literature shows an increasing amount of interesting studies on direct microbial electron uptake from cathodes by autotrophic organisms, in which a cathodic hydrogen production can be vastly excluded by means of thermodynamics. Still, these systems are so far limited by a number of factors, including for instance a comparatively low current consumption [18], a dependence on enrichment cultures [19,20,21], or the necessity to provide light as a further energy source to the microorganisms [22, 23].
Although there are many CO2 assimilating cathodic communities described, the literature only knows a handful of microorganisms that are suitable as a pure culture biocatalyst for MES [20, 21, 24,25,26,27,28]. So far, most of these organisms belong to the physiological group of either methanogens or acetogens, resulting in a rather small product range in MES [6, 8, 12, 13, 29,30,31]. The production of more complex compounds by these organisms is hampered by the energy limitation that is combined with the use of carbon dioxide as electron acceptor [32]. Therefore, the production of more complex compounds or the genetic engineering aiming at the production of new compounds is at least challenging with biocatalysts that are dependent on the Wood–Ljungdahl pathway [33, 34]. Large CO2 emission sources such as refineries or power plants typically exhaust flue gas that exceeds the optimum growth temperature of mesophilic microorganisms and typically contain oxygen. The application of oxygen tolerant thermophilic biocatalysts in a MES process could avoid the need of a time and energy consuming exhaust gas cooling and purification. Moreover, extreme culturing conditions will lower the risk of contamination in a future biotechnological process.
In this study, we present the enrichment and characterization of the first thermoacidophilic, electroautotrophic community. In this regard, we see electroautotrophic growth characterized by (I) the electrode being directly or indirectly the electron and energy donor of the organisms, (II) a measurable and to the activity of the organisms assignable change of the cathode reaction, and (III) physical contact of the organisms with the electrode surface. We isolated distinct species from the initial cathodic enrichment. One of the isolates belongs to the physiological group of Knallgas bacteria. This organism is a natural producer of polyhydroxybutyrate (PHB), a microbial storage compound that gained increased attention as a bioplastic precursor [35,36,37]. Our data reveals, that the novel isolate has the potential to serve as a biocatalyst in an oxic, thermoacidophilic MES process. This would combine the benefit of a sustainable bioproduction with an easily recyclable biopolymer as end product.
Results
Initial enrichment of the electroautotrophic community
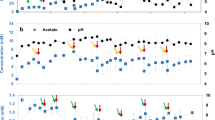
Samples from geothermal hot springs on the Azores were used to inoculate microbial electrochemical cells. The single chamber reactors contained a 16 cm2 carbon felt cathode and a platinum net anode separated from the medium filled reactor by a glass frit. The applied minimal salt medium (ES medium, pH 3.5) was developed according to the hydrogeochemistry of the sampling site [38]. The potential of the working electrode was set to −381 mV vs. standard hydrogen electrode (SHE) (−594 mV vs. saturated calomel electrode). During the incubation of the sediment samples at 60 °C, a gradual increase of the reductive current was observed. To eliminate remaining organics carried over from the inoculum and to force a selection for electroautotrophic biofilm communities, the cathode was transferred to a sterile reactor with sterile medium four times within the enrichment period of 18 months. The development of a stable reductive current was used as indication that the community stabilized and the medium could be exchanged. After the last transfer, the reductive current (expressed as negative current) in the inoculated reactor increased to −80 µA cm−2, while the current of the sterile control was stable at roughly −10 µA cm−2 over a period of 90 days (Fig. 1a). Microscopic analysis was conducted to confirm that the observed current was due to microbial activity (Fig. 1 and Supplementary Fig. 1). CARD-FISH analysis of the cathode graphite fibers revealed the presence of a biofilm that was dominated by bacteria, while archaea were detected only to a minor extend (Supplementary Fig. 1). The morphology of the biofilm was further investigated by scanning electron microscopic analysis. The electron micrographs showed cell agglomerates between and around the graphite fibers (Supplementary Fig. 1A, B). This result was corroborated by fluorescence microscopy data (Supplementary Fig. 1D, F). Autotrophic growth of the initial enrichment was measured via the incorporation of 13C-labeled bicarbonate. To that end, the biofilm coated electrode was cut in half and placed in the same reactor but only one of the electrodes was poised to −381 mV vs. SHE, while the other half was not connected to the potentiostat (Fig. 1b). The sample gained from the poised cathode revealed a δ13C of 61‰ after 1 week and 226‰ after 7 weeks, compared with −28‰ and −20‰ for the unconnected cathode. Hence, the experiment showed that δ13C shifted only to positive values in the samples that were supplied with electrons from the cathode surface, which indicates current based autotrophic activity. HPLC analyses of both, the reactor medium and the cold trapped gas efflux, revealed no accumulation of organic compounds, which suggests carbon cycling within the thermoacidophilic biofilm community.
a Current density of the initial enrichment reactor (red) and a sterile control (black) over time. Cathodes of both reactors were poised to −381 mV vs. SHE. The observed current density fluctuations of the inoculated system correlate with variations in the gas flow (N2/CO2, 80:20, v/v) through the reactor and the compensation of media evaporation by the addition of sterile ddH2O. b Experimental timeline of the 13C-incoporation experiment. The table contains the measured δ13C isotopic signatures in parts per thousand [‰]. An integration of the 13C-labeled carbonate is characterized by a positive δ13C value.
Metagenomic analyses of the initial enrichment
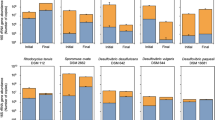
A metagenome of the community was assembled to assess its physiological capabilities. Of note, due to limited availability of cathode material, we had to amplify the nucleic acids isolated from the biofilm using isothermal amplification. Phylogenetic binning was conducted either by using the blast algorithm on the individual genes of assembled contigs (one contig was assigned to only one phylogenetic group) or the MetaBat-work flow published recently [39]. The results are similar but the quantitative assignment of DNA sequences to individual phylogenic groups differed. The Blast-based analysis revealed that the three most abundant bacterial families belong to the Firmicutes: more than 55% of all deduced protein sequences grouped within the families Thermoanaerobacteraceae (35.6%), Peptococcaceae (21.7%), and Sporomusaceae (1.8%). 18.6% of all sequences were attributed to six groups within the archaea (Fig. 2a and Supplementary Table 1). Within the Thermoanaerobacteraceae—Moorella, Caldanaerobacter and Thermodesulfitimonas are the most abundant genera. Desulfofundulus is the main genus within the Peptococcaceae. The main genera of the archaea seem to be Sulfolobus, Thermoplasma, and Acidianus. For the MetaBat-analysis 29,797 contigs were obtained ranging in length between 200 and 331,073 bp. Those contigs were ranked into 52 bins. With Bowtie2 we could assign 69% of all reads to one of 52 bins (Fig. 2b and Supplementary Table 2). Although, all above listed genera could also be detected as phylogenetic bin using this analysis, Thermodesulfitimonas, Sulfolobus, and Aciduliprofundum seem to be overrepresented using this analysis.
a Taxonomic distribution, corresponding to annotated proteins in the metagenome (Only taxa with an abundancy >0.2% are depicted; other: unassigned genes and underrepresented taxa with an abundancy ≤0.2%; displaced wedges represent archaea). b Taxonomic distribution based on the conducted MetaBat analysis. Filtered raw reads of the metagenome were mapped against the 52 phylogenetic bins. Stacked bars of the asterixed taxa consist of several bins with identical taxonomic contribution. The full summary of all bins can be found in Supplementary Table 2.
The metagenome was screened for the presence of genes encoding carbon dioxide fixation pathways, potential pathways for electron uptake from the cathode and terminal reductases. Genes encoding key enzymes for the Wood–Ljungdahl pathway, the reverse citric acid cycle, the 3-hydroxypropionate/4-hydroxybutyrate pathway, the dicarboxylate/4-hydroxybutyrate pathway as well as the Calvin cycle were detected using the KEGG database as well as a specifically designed database (Supplementary Table 8) comprising sequences of key enzymes (Supplementary Table 3). Within the dominant phylogenetic bins we identified genes encoding for key enzymes of the Wood–Ljungdahl pathway in the acetogenic genera Moorella, Desulfofundulus, and Thermodesulfitimonas, whereas carbon fixation via the dicarboxylate/4-hydroxybutyrate and the 3-hydroxypropionate/4-hydroxybutyrate cycle was confirmed for Sulfolobus, Acidianus, and Metallosphaera (Supplementary Tables 3 and 4).
Regarding potential electron uptake mechanisms, it was sought for hydrogenases, formate dehydrogenases as well as enzymes used by ferrous iron oxidizing microorganisms. The latter were identified using a specific database containing sequences of the key electron transferring enzymes like rusticyanin and fox homologs (Supplementary Table 9). Moreover, the database contained sequences of enzymes (MtrABC) involved in extracellular electron transfer (EET) of dissimilatory iron reducers like Shewanella oneidensis, as studies suggest that these organisms can also mediate a reversed electron transfer from a cathode into the periplasm [40,41,42]. In addition, the database included sequences of pioABC and mtoABD gene clusters as the corresponding proteins catalyze an extracellular electron uptake during iron oxidation in Rhodopseudomonas palustris and Sideroxydans lithotrophicus. The bioinformatic analysis revealed the presence of genes coding for homologous proteins to PioA, MtoA, MtrAB, rusticyanins, and the Fox proteins (e value < 0.001; Supplementary Table 5). However, a full set of homologous genes, encoding for a theoretically functional Mtr/PioABC complex, was not found within the metagenome. Homologs sequences to rusticyanin and fox proteins were identified but a direct interaction of these proteins with electrodes was so far not elucidated. In contrast, the occurrence of hydrogenases and formate dehydrogenases was confirmed for all dominant phylogenetic bins (with the exception of the genera Acidianus and Caldanaerobacter that only contained hydrogenases), indicating a hydrogen or formate mediated primary electron uptake. Still, it is not clear whether the detected hydrogenases are involved (I) in the uptake of hydrogen that formed abiotically at the electrode surface, (II) in the direct catalyzes of hydrogen production, or (III) the direct import of electrons into the cytoplasmic membrane of the organisms. Regarding potential electron acceptors that could be used by the key genera, we identified the genes necessary for carbonate respiration (Moorella, Desulfofundulus, and Thermodesulfitimonas), as well as sulfate (Desulfofundulus) and oxygen (Acidianus, Metallosphaera, and Sulfolobus) reduction. Since the enrichment reactor was a membrane-less bioelectrochemical system, low amounts of anodically produced oxygen could possibly close an interspecies sulfur cycle between the Crenarchaeota and Desulfofundulus. Thermodesulfitimonas, the most abundant organism according to the MetaBat analysis was described to thrive using sulfite as electron acceptor and hydrogen as electron donor and consequently could also be part of the postulated sulfur cycle [43]. Moreover, Aciduliprofundum, the second most abundant genus according to MetaBat, is a heterotrophic sulfur and ferric iron reducing organism and would be a potential secondary organism within the community. Since the cathodic community is dominated by obligate anaerobic bacteria (e.g., Moorella, Desulfotomaculum, and Thermodesulfitimonas) the planctonic oxygen reduction seems to be sufficient to maintain anoxic conditions at the cathode. Moreover, the underrepresented Knallgas bacteria of the genus Kyrpidia could serve as an additional oxygen scavenger in the cathodic biofilm. Based on the HPLC results and the metagenomic analyses, Fig. 3 depicts a scheme of the potential metabolic interaction within the cathodic biofilm.
Electrons could be taken up from the cathode either directly or via hydrogen or formate dehydrogenase. These electrons could provide the energy source for Desulfofundulus species and organisms belonging to the Thermoanaerobacteraceae. The latter would produce acetate as catabolic end product which could be used by a variety of heterotrophic species in the bioelectrochemical system. Hydrogen sulfide provided by Desulfofundulus or Thermodesulfitimonas species reacts abiotically with divalent metal cations providing an electron donor for crenarchaeotal species. Oxygen originating from the anode reaction of the system could be the electron acceptor for these organisms. Carbon dioxide fixation pathways discovered in the bioinformatic analysis are indicated by WL (Wood–Ljungdahl-pathway), 3HP/4HP DC/4HB (3-hydroxypropionate/4-hydroxybutyrate or dicarboxylate/4-hydroxybutyrate cycle), and CC (Calvin cycle).
Isolation of distinct species from the initial enrichment
In order to screen for new biocatalysts suitable for a thermophilic electrosynthesis (ES) process, we tried to isolate potential electroautotrophic species from the initial enrichment. Five isolates were obtained that belong to the genera Thermoanaerobacterium, Desulfofundulus, Acidianus, Alicyclobacillus, and Kyrpidia. The isolates are most closely related to: Thermoanaerobacterium saccharolyticum, Desulfofundulus thermobenzoicus, Acidianus brierleyi, Alicyclobacillus acidocaldarius, and Kyrpidia tusciae (please find the 16S rRNA sequences of the respective organisms in Supplementary Table 6). The Kyrpidia isolate was gained using hydrogen as electron donor and oxygen as electron acceptor and was found to be a novel, undescribed species, which was named Kyrpidia spormannii and characterized with regards to its phylogenetic position in a previous study [44]. Thermoanaerobacterium sp., Desulfofundulus sp., and Acidianus sp. are representatives of the three most abundant families of the initial cathodic biofilm. All isolates were tested for lithoautotrophic growth in adequate media (See Supplementary Material and Methods). As no lithoautotrophic growth could be observed in experiments with Thermoanaerobacterium sp. and Alicyclobacillus sp., both isolates were not used in the subsequent screening. Finally, the ability of the remaining isolates to grow on cathodes was assessed in a bioelectrochemical system using carbon dioxide as sole carbon source. The same media as employed for lithoautotrohic growth were used in these experiments (without added electron donor). The applied potential of the working electrode was poised to −531 mV vs. SHE (−744 mV vs. saturated calomel electrode) to accelerate growth of the organisms. Reactors inoculated with a pure culture of Desulfofundulus sp. or Acidianus sp. showed no significant current consumption compared with sterile control reactors. However, the inoculation of ES reactors with a pure culture of Kyrpidia spormannii revealed a significant effect in cathodic current consumption (Fig. 4a). Overall, 0.3% of all genes in the metagenome could be assigned to the genus Kyrpidia (Fig. 2a).
a Chronoamperometric data comparing inoculated electrosynthesis reactors (red) with an abiotic control (black). Cathodes were poised to −531 mV vs. SHE. The sparging gas had an oxygen concentration of 0.5%. Gray error bars represent standard deviation (n = 3). b Representative electron micrographs of the graphite electrodes after 9 days of incubation in an electrosynthesis reactor inoculated with K. spormannii. A cathode that was poised to −531 mV vs. SHE is depicted on the left-hand side, whereas the right picture shows an open circuit potential (OCP) control cathode.
Electroautotrophic experiments with Kyrpidia spormannii
As K. spormannii is described as an obligate aerobic organism, the bioelectrochemical reactors were constantly sparged with a gas mixture of 99.5% CO2 and 0.5% O2 (v/v) [44]. After an initial polarization time of 2 days, the current density of a sterile control reactor stabilized at approximately −15 µA cm−2. By contrast, the current density of an inoculated reactor dropped down to −50 µA cm−2 over a period of 10 days (Fig. 4a). This effect was not observed in a reactor inoculated with the most closely related isolate K. tusciae DSM 2912 (Supplementary Fig. 2). Scanning electron micrographs of cathodes taken after 9 days of incubation revealed a higher cell density on an electrode poised to a potential of −531 mV vs. SHE compared with an open circuit potential (OCP) control (Fig. 4b).
Comparative transcriptomic analyses of Kyrpidia spormannii
To proof an autotrophic lifestyle of K. spormannii growing on a cathode and to reveal possible electron uptake mechanisms, comparative transcriptomic analyses were conducted. The expression profile of cells growing as biofilm on a cathode (−531 mV vs. SHE) was compared with heterotrophically and lithoautotrophically (hydrogen as electron donor) grown cells. The principal component analysis of the obtained transcriptomic data revealed that the different growth conditions led to distinct expression profiles (Supplementary Fig. 3). Analysis of the previously sequenced genome of K. spormannii, indicated the presence of genes encoding for the Calvin cycle [45]. The expression of these genes was negligible under heterotrophic conditions, whereas a strong expression of the Calvin cycle was detected under lithoautotrophic and electroautotrophic growth conditions (Fig. 5). As the fixation of carbon dioxide is dependent on reducing equivalents, genomic and transcriptomic data were used to create a model of the respiratory chain of K. spormannii to follow the route of NADPH and ATP generation (summarized in Fig. 6). As no genes showed a significant homology to any known NAD(P) transhydrogenase gene, other possibilities of NADPH generation were investigated. The transcriptomic data revealed a ferredoxin dependent NADP reductase that was upregulated under electro- and lithoautrotrophic conditions. Furthermore, we found two distinct gene clusters coding for two Nuo-complexes. Interestingly, both complexes consist of 11 subunits lacking the electron input module nuoE–nuoG. The absence of those subunits, involved in the binding of NADH in Escherichia coli, can also be observed in cyanobacteria, chloroplasts, and most SRBs [46, 47]. Therefore, a specific direct interaction with NAD(P)H seems to be unlikely. Yet, ferredoxin was suggested as a possible substrate of the enzyme complex [48]. The first cluster (complex Ia) was only expressed under litho- and electroautotrophic conditions, whereas the second complex (Ib) was expressed under all three conditions, showing a slight downregulation under electroautotrophic conditions. Moreover, the genes of the Calvin cycle, the Nuo-complex Ia, as well as the ferredoxin:NADP reductase are located in the same cluster controlled by the Calvin cycle regulator CbbR [49]. This leads to the assumption, that the Ia complex is involved in NADPH generation via a proton motive force driven reversed electron flow, oxidizing the menaquinone pool of K. spormannii (MK-7) [44]. The transcriptomic data were inspected for potential electron uptake mechanisms, replenishing the quinone pool. So far, most direct electron uptake from solid surfaces is known to be conducted via (multiheme) c-type cytochromes [50]. Based on the heme-binding peptide motif CXXCH, 34 c-type cytochromes were identified in the genome of K. spormannii (33 monoheme and one diheme cytochrome) of which two genes were significantly upregulated (p < 0.01; log2-fold change > 2) under electroautotrophic growth compared with autotrophic and heterotrophic growth, respectively [51]. However, the absolute expression of these two cytochromes is rather low (Table 1). Furthermore, the expression of the [NiFe]-hydrogenase and formate dehydrogenases, as potential catalysts for an indirect electron uptake mechanism, was investigated. No significant differences in the expression of formate dehydrogenases were observed. The hydrogenase subunits were upregulated significantly (p < 0.01; log2-fold change > 3.5) under lithoautotrophic and electroautotrophic conditions. In the latter case, both subunits belonged to the 40 most expressed genes. The in silico analyses did not resolve the question, if this uptake hydrogenase transfers the electrons directly or indirectly into the menaquinone pool. In order to complete the respiratory chain, it was sought for genes encoding menaquinol oxidases. The bioinformatic screen resulted in the identification of a branched respiratory chain in K. spormannii. From the menaquinone pool, oxygen can be reduced via the classical cytochrome c pathway involving the respective oxidoreductases (complex III and IV) or directly via two different menaquinol oxidases (cytochrome bd and cytochrome aa3-600 type). Whereas the expression of the menaquinone:cytochrome c oxidoreductase (complex III) is downregulated under electroautotrophic conditions, the two menaquinol oxidases are upregulated during cathodic growth. Only a minor change in the expression of the F-type ATP synthase was observed (Fig. 6b).
Participating genes (gene number) and the corresponding relative expression changes (log2FC log2-fold change) are depicted in boxes. Gene copies without a pertinent expression were omitted (concerned enzymes are marked with an asterisk). TKT transketolase, FBP fructose-1,6-bisphosphatase, FSA fructose-6-phosphate aldolase, FBA fructose-1,6-bisphosphate aldolase, RPIB ribose-5-phosphate isomerase, PRK phosphoribulokinase, RuBisCO ribulose-bisphosphate carboxylase/oxygenase, GAPDH glyceraldehyde-3-phosphate dehydrogenase, PGM phosphoglycerate mutase, ENO enolase, PK pyruvate kinase, PDH pyruvate dehydrogenase, PFOR pyruvate-ferredoxin oxidoreductase, ACAT acetyl-CoA acyltransferase, PhaB acetoacetyl-CoA reductase, and PhaC PHA polymerase.
Compared with lithoautotrophic conditions, a total of 323 genes were significantly upregulated (p < 0.01; log2-fold-change > 2) under electroautotrophic conditions, from which 129 genes were annotated as hypothetical proteins. The three most significantly upregulated genes with an annotated function were genes encoding for a nucleotide sugar dehydrogenase, a spore-coat protein, and hemerythrin. As K. spormannii is described as PHB producing organism, the transcription of genes contributing to a putative pathway for the production of PHB from CO2 was investigated (Fig. 5). Transcripts corresponding to all necessary enzymes for the conversion of carbon dioxide to PHB were detected. Moreover, expression of phaB and phaC encoding the acetoacetyl-CoA reductase and PHA polymerase was increased compared with lithoautotrophic or heterotrophic growth (Fig. 5). Still, the p values show significant upregulation only from lithotrophic to electroautotrophic growth conditions. The full expression table including comparative statistical information and amino acid sequences of the corresponding genes can be found in the supplement (Supplementary Table 7).
Flow-cell experiments
So far, we were able to measure metabolic activity of cathodically grown K. spormannii cells but the results did not allow to conclusively correlate metabolic activity with growth. Hence, we established a platform to study cathodic biofilm growth in situ. By providing a pressurizable flow-cell setup, this engineering effort was combined with a simultaneous increase of the dissolved CO2 and oxygen concentration. The transparent glass lid allowed noninvasive imaging of biofilm growth on the cathode by means of Optical coherence tomography (OCT) (Supplementary Fig. 4). The biofilm formation on a cathode poised to −531 mV vs. SHE (Fig. 7a–c) was compared with the biofilm formation on an OCP control cathode (Fig. 7e). A noninoculated reactor served as an additional control (−531 mV vs. SHE) (Fig. 7d). After 10 days of incubation at 60 °C, both control cathodes were only partly covered by a precipitate-like structure, whereas the poised cathode from the inoculated system was confluently covered by a clearly three-dimensional biofilm-like structure with an average height of 80 µm (Fig. 7). Microscopic analysis of the biofilm showed rod-shaped cells covered in an unidentified matrix compound (Supplementary Fig. 5). Confocal microscopy of Nile red stained biofilm samples revealed distinct PHA inclusions (Fig. 7f). An aggregation of cells at the cathode surface due to sedimentation was rather unlikely since a rather high flow rate (100 ml min−1) was used and no electrostatic effect was expected since the cathode and the bacterial surface are both negatively charged. Yet, to corroborate cathodic carbon dioxide fixation of the isolate, we inoculated a flow cell with a “13C-loaded” K. spormannii culture (85.3 ± 0.6 at% of total carbon was 13C labeled). After 10 days of incubation offering unlabeled CO2 as the sole carbon source, the total carbon of the harvested biofilm contained only 36.6 ± 4.7 at% 13C, indicating an active incorporation of the supplied 12CO2 under electroautotrophic conditions. By employing this reversed labeling approach, an extensive consume of costly 13C labeled CO2 during the flow-cell operation was avoided.
Discussion
In this study, we present an experimental workflow following the principle of Robert Koch’s postulates ranging from the observation of an effect in a laboratory ecosystem to the isolation of individual species and the verification of an activity that was also observed in the multispecies ecosystem. This is the first isolation of an electroautotrophic microorganism from a mixed species cathodic biofilm that was enriched from environmental samples. Although K. spormannii was underrepresented in the initial cathodic enrichment, the physiological capabilities of the isolate resemble the chosen enrichment conditions and, thus showing the benefit that isolation campaigns can have. Almost all so far reported studies on MES were performed under moderate conditions such as neutral pH and mesophilic temperatures [12, 14, 52,53,54]. We describe the first thermoacidophilic, electroautotrophic consortium. The reductive current densities that were obtained during the enrichment phase are higher than values typically measured in previous studies conducted with a similar cathode potential but other electroautotrophic biocatalysts. Nevertheless, the comparability even of the so far published studies is low since cathode material, temperature, pH, and reactor configuration influence the bioelectrochemical measurements [55]. Moreover, in the here described experiments a one chamber setup was used. This could, at least to some extent, lead to an abiotic oxygen reduction at the cathode surface.
According to CARD-FISH analysis, the cathodic biofilm was dominated by bacteria. This is corroborated by the metagenomic data, since only 18.6% of the deduced protein sequences could be phylogenetically assigned to archaea using the conducted blast analysis of the genes on the assembled contigs (each contig was assigned to one taxa). Similarly, 29.8% of the normalized reads have mapped to archaeal bins that where assembled from the MetaBat-analysis, respectively. The most dominant bacterial family of the initial enrichment are the Thermoanaerobacteraceae. Nevin et al. applied Moorella thermoacetica, a member of the Thermoanaerobacteraceae, to produce acetate using cathodic electrons at −400 mV vs. SHE as electron donor [12, 56]. The second most abundant family Peptococcaceae includes autotrophic SRBs such as Desulfosporosinus and Desulfofundulus [57, 58]. This physiological group seems to be involved in microbial iron corrosion and has the ability for cathodic electron uptake and hydrogen production [58,59,60,61,62]. Finally, the third most abundant bacterial family Sporomusaceae comprises the species Sporomusa ovata, a well described biocatalyst for MES [6, 30, 63]. Looking at the archaeal community, the applied conditions led to an enrichment of organisms belonging to the family of Sulfolobaceae.
Evidence for cathode-driven autotrophy comes from an increase in δ13C that was only observed in cells obtained from a poised cathode. The specific isotopic fractionation seen in the unpoised samples corroborates the metagenomic data, that the Wood–Ljungdahl pathway and the 4-hydroxybutyrate pathways mainly contribute to microbial CO2 fixation [33]. Taken together, this indicates that the members of the most abundant families in the community could be involved in electroautotrophic primary production.
We further assessed the metagenome to identify possible entry points for cathodic electrons. A conventional c-type cytochrome-based direct electron uptake as the main entry point for electrons was found to be rather unlikely, since the genetic information of the designated key species contained, if any, only genes coding for an incomplete electron transfer complex. By contrast, hydrogenases were found for all of the most common genera, indicating a hydrogenase-based electron uptake. However, the metagenomic analysis cannot resolve the actual role of the hydrogenases during cathodic electron uptake. At the chosen working potentials, the possibility of an abiotic or hydrogenase mediated cathodic hydrogen generation cannot be ruled out. A hydrogen production by a Desulfofundulus species is also conceivable, since a direct electron uptake leading to the formation of molecular hydrogen as intermediate was often postulated for iron-corroding SRBs. Still, the isolated Desulfofundulus strain did not show an increased current consumption in an ES reactor compared with a sterile control. This leads to the hypothesis that this strain might thrive in the biofilm using acetate generated by the acetogenic species as an electron source for respiratory sulfate reduction or hydrogen production since the minimal medium contained about 1 mM of sulfate [64]. A similar type of interaction between an acetogenic organism and a SRB was also postulated in another study describing a mesophilic cathodic enrichment conducted at a cathodic potential of −590 mV vs. SHE [65]. The oxygen dependent crenarchaeota could use reduced sulfur species as an electron source thereby closing the sulfur cycle. Intermediates of the sulfur cycle are further necessary to support growth of Thermodesulfitimonas and Aciduliprofundum species that thrive using sulfite or sulfur as electron acceptor. So far, we cannot say what the major source of both sulfur species might be.
K. spormannii was isolated under microaerophilic conditions and is one of the first electroautotrophs not belonging to the physiological group of methanogens or acetogens [44]. Even though the initial enrichment was conducted under an N2/CO2 atmosphere, small but constant amounts of oxygen were formed at the anode. This oxygen limitation could be the reason why this species was underrepresented in the initial electroautotrophic enrichment. Nevertheless, 0.3% of all genes in the metagenome were assigned to the genus Kyrpidia. Hence, these organisms will have found sufficient amounts of electron acceptor to thrive in the cathodic biofilm. Moreover, Knallgas bacteria based oxygen depletion within the cathodic biofilm could have provided other strictly anoxic organisms of the community with a growth advantage.
The expression profile of K. spormannii cells grown on a cathode surface not only provides the first transcriptomic data of an electroautotrophic growing pure culture, it also shows a distinct regulatory response to electroautotrophic growth (Supplementary Fig. 3). As the cells were growing sessile, the expression of genes, like nucleotide sugar dehydrogenases, that could be involved in biofilm and EPS (extracellular polymeric substances) formation, were upregulated under electroautotrophic conditions [66]. In addition, the transcriptomic data revealed a highly expressed Calvin cycle under electroautrotrophic conditions. The expression of this energetically demanding pathway and the result of the 13C labeling experiment proves the autotrophic lifestyle of the cathodic biofilm [67].
Based on the transcriptomic data, a model of the branched respiratory chain of K. spormannii was established. We found two homologous clusters encoding for two type I NADH dehydrogenase complexes, putatively one with an anabolic and the other with a reversed, catabolic activity. Noteworthy, the nuo gene clusters lack the genes nuoE–nuoG. These genes are known to encode for the NADH binding domain of complex I, leading to the hypothesis of the involvement of an alternative substrate [68]. The expression changes in the different menaquinone oxidase modules during electroautotrophic conditions could be an effect of the adaption to an oxygen and nutrient limitation in the biofilm, or various stresses like reactive oxygen species. Especially cytochrome bd type menaquinol oxidases are known to show a high oxygen affinity [69]. To find a potential entry point for the cathodic electrons into the respiratory chain of K. spormannii, we took different electron uptake mechanisms into consideration:
In most microorganisms EET is mediated by c-type cytochromes. With exemptions, this mechanism seems to be specifically employed by Gram-negative bacteria [70]. No highly upregulated c-type cytochromes were detected under electroautotrophic conditions in K. spormannii that was described to be Gram positive. Hence, electron uptake seems to differ compared with Candidatus “Tenderia electrophaga” for which evidence was provided that electron import might occur via the catalytic activity of cytochromes. Of note, this organism can thrive at cathodes even poised to 0.31 mV versus SHE [21]. Transcriptomic analysis of cathode grown K. spormannii also revealed strong upregulation of a di-iron hemerythrin. If this protein has a role in EET or in its so far known function as oxygen carrier, remains unclear [71]. Interestingly, BLAST analysis revealed that the hydrogenase of K. spormannii shares a comparatively low protein identity with the hydrogenase of the phylogenetically closest relative K. tusciae [44] (Supplementary Figs. 6–8). Both hydrogenase subunits were found to be highly expressed under electroautotrophic conditions, making the assumption of a hydrogenase dependent electron uptake to be most likely. Moreover, the highest protein identity of the electron transferring small hydrogenase subunit is shared with that of Acidithiobacillus ferrooxidans [72]. At pH 2, this aerobic iron oxidizer takes up cathodic electrons with a potential of 0 mV vs. SCE [73]. The literature reports highly differing current densities that can be obtained with Acidithiobacillus strains ([18, 73, 74]) which might be due to the above mentioned potential abiotic oxygen reduction on the cathode surface. Still, the cathode potential in all studies dealing with Acidithiobacillus strains was higher compared with our study which might suggest direct rather than indirect interaction with the cathode surface.
So far, no other Knallgas bacterium is reported to grow on cathodes as sole energy and electron source at potentials as high as −531 mV vs. SHE [75, 76]. In fact, another study using Cupriavidus necator as catalyst in the cathode compartment of a bioelectrochemical system displayed that the ability to grow lithoautotrophically alone is not sufficient for biofilm growth on cathode surfaces most probably due to the formation of reactive oxygen species [17]. A confluently biofilm covered cathode could be a strategy to cope with oxidative stress as oxygen can be depleted within the biofilm before it reaches the electrode surface. Our data indicates a potential biotechnological application for K. spormannii in the field of MES. K. spormannii is a natural producer of polyhydroxyalkanoates which are biopolymers that have already been established as consumer products. Still, they are so far produced under heterotrophic conditions which drastically lowers the production efficiency and interferes with the usage of crops for the food sector.
Methods
Electrochemical culturing conditions of the initial enrichment
Environmental samples were collected in August 2012 from hydrothermal systems on São Miguel Island (Azores, Portugal). The GPS coordinates of the sampling site are published elsewhere [44]. The initial enrichment reactor was inoculated with a mixture of 24 water and sediment samples. Chronoamperometry was performed in a reactor that was described previously [77]. The working electrode chamber volume was 2 l. A 16 cm2 piece of graphite felt (SGL Carbon SE, Wiesbaden, Germany) served as working electrode (cathode). A platinum gauze (0.5 × 5 cm, 1024 mesh cm−2, wire 0.06 mm diameter), inserted into a porous glass tube, was used as counter electrode (anode). As reference electrode, a saturated calomel electrode (SCE) (Sensortechnik Meinsberg, Waldheim, Germany) was employed. For the initial enrichment, an ES minimal medium was used [44]. The pH was adjusted to 3.5 with H2SO4. During the total incubation time of 18 months, the medium was exchanged carefully several times, avoiding a disruption of the cathodic biofilm. The reactor was connected to a Multi Autolab M101 potentiostat (Metrohm, Herisau, Switzerland) and a constant potential of −594 mV vs. SCE was applied. The system was incubated at 60 °C and continuously sparged with a mixture of N2/CO2 (80:20, v/v). Metagenomic and CARD-FISH analyses were carried out after 18 months of incubation. Implementation of the 13C-bicarbonate fixation experiment and microscopic analyses is available in the Supplementary Material and Methods section online.
Isolation of distinct species from initial cathodic enrichment
For enrichment media and conditions see Supplementary Material and Methods.
Evaluation of electroautotrophic growth with Kyrpidia spormannii
Electroautotrophic growth of Kyrpidia spormannii was tested at 60 °C in the same 2 l reactors described before [77]. ES medium (pH 3.5) was used and a constant potential of −531 vs. SHE was applied for at least 10 days. The system was constantly sparged with a mixture of 99.5% CO2 and 0.5% O2 (v/v). The system was inoculated with 20 ml of a mid-log phase, lithoautotrophic culture [44]. The control experiments with K. tusciae were performed under same conditions. Noninoculated reactors served as negative controls. Chronoamperometric experiments where performed in triplicates.
Nucleic acid isolation and amplification, Illumina sequencing, and bioinformatic analyses
See Supplementary Material and Methods for metagenomic analysis conducted with the initial enrichment, as well as the comparative transcriptomic analysis of K. spormannii.
Flow-cell experiments
See Supplementary Material and Methods for further details on flow-cell operation.
Data availability
Data supporting the findings of this study are available within the article and its Supplementary Information Files and from the corresponding author upon reasonable request. All raw sequencing data can be found on NCBI short read archive (PRJNA483115 and PRJNA477433).
References
Pachauri RK, Allen MR, Barros VR, Broome J, Cramer W, Christ R, et al. Climate change 2014: synthesis report. Contribution of Working Groups I, II and III to the fifth assessment report of the Intergovernmental Panel on Climate Change. Geneva, Switzerland: IPCC; 2014.
Lieder M, Rashid A. Towards circular economy implementation: a comprehensive review in context of manufacturing industry. J Clean Prod. 2016;115:36–51.
European Commission. Press release—closing the loop: commission adopts ambitious new circular economy package to boost competitiveness, create jobs and generate sustainable growth. Brüssel: New Europe; 2015.
UNFCCC. Conference of the Parties. Report of the Conference of the Parties on its twenty-first session, held in Paris from 30 November to 13 December 2015. Paris, France: UNFCCC; 2015.
Cheng S, Xing D, Call DF, Logan BE. Direct biological conversion of electrical current into methane by electromethanogenesis. Environ Sci Technol. 2009;43:3953–8.
Nevin KP, Woodard TL, Franks AE, Summers ZM, Lovley DR. Microbial electrosynthesis: feeding microbes electricity to convert carbon dioxide and water to multicarbon extracellular organic compounds. MBio. 2010;1:e00103–10.
Sadhukhan J, Lloyd JR, Scott K, Premier GC, Yu EH, Curtis T, et al. A critical review of integration analysis of microbial electrosynthesis (MES) systems with waste biorefineries for the production of biofuel and chemical from reuse of CO2. Renew Sustain Energy Rev. 2016;56:116–32.
Rosenbaum MA, Franks AE. Microbial catalysis in bioelectrochemical technologies: status quo, challenges and perspectives. Appl Microbiol Biotechnol. 2014;98:509–18.
Christodoulou X, Okoroafor T, Parry S, Velasquez-Orta SB. The use of carbon dioxide in microbial electrosynthesis: Advancements, sustainability and economic feasibility. J CO2 Util. 2017;18:390–9.
Butler CS, Lovley DR. How to sustainably feed a microbe: strategies for biological production of carbon-based commodities with renewable electricity. Front Microbiol. 2016;7:1879.
Igarashi K, Kato S. Extracellular electron transfer in acetogenic bacteria and its application for conversion of carbon dioxide into organic compounds. Appl Microbiol Biotechnol. 2017;101:6301–7.
Nevin KP, Hensley SA, Franks AE, Summers ZM, Ou J, Woodard TL, et al. Electrosynthesis of organic compounds from carbon dioxide is catalyzed by a diversity of acetogenic microorganisms. Appl Environ Microbiol. 2011;77:2882–6.
Deutzmann JS, Sahin M, Spormann AM. Extracellular enzymes facilitate electron uptake in biocorrosion and bioelectrosynthesis. MBio. 2015;6:1–8.
Lohner ST, Deutzmann JS, Logan BE, Leigh J, Spormann AM. Hydrogenase-independent uptake and metabolism of electrons by the archaeon Methanococcus maripaludis. ISME J. 2014;8:1673–81.
Lovley DR, Nevin KP. Electrobiocommodities: powering microbial production of fuels and commodity chemicals from carbon dioxide with electricity. Curr Opin Biotechnol. 2013;24:385–90.
Beese-Vasbender PF, Grote J-P, Garrelfs J, Stratmann M, Mayrhofer KJJ. Selective microbial electrosynthesis of methane by a pure culture of a marine lithoautotrophic archaeon. Bioelectrochemistry. 2015;102:50–5.
Li H, Opgenorth PH, Wernick DG, Rogers S, Wu TY, Higashide W, et al. Integrated electromicrobial conversion of CO2 to higher alcohols. Science. 2012;335:1596.
Ishii T, Kawaichi S, Nakagawa H, Hashimoto K, Nakamura R. From chemolithoautotrophs to electrolithoautotrophs: CO2 fixation by Fe(II)-oxidizing bacteria coupled with direct uptake of electrons from solid electron sources. Front Microbiol. 2015;6:994.
Eddie BJ, Wang Z, Malanoski AP, Hall RJ, Oh SD, Heiner C, et al. ‘Candidatus Tenderia electrophaga’, an uncultivated electroautotroph from a biocathode enrichment. Int J Syst Evol Microbiol. 2016;66:2178–85.
Malanoski AP, Lin B, Eddie BJ, Wang Z, Hervey WJ, Glaven SM. Relative abundance of ‘Candidatus Tenderia electrophaga’ is linked to cathodic current in an aerobic biocathode community. Microb Biotechnol. 2018;11:98–111.
Eddie BJ, Wang Z, Hervey WJ, Leary DH, Malanoski AP, Tender LM, et al. Metatranscriptomics supports the mechanism for biocathode electroautotrophy by “Candidatus Tenderia electrophaga”. mSystems. 2017;2:e00002–17.
Guzman MS, Rengasamy K, Binkley MM, Jones C, Ranaivoarisoa TO, Singh R, et al. Phototrophic extracellular electron uptake is linked to carbon dioxide fixation in the bacterium Rhodopseudomonas palustris. Nat Commun. 2019;10:1355.
Bose A, Gardel EJ, Vidoudez C, Parra EA, Girguis PR. Electron uptake by iron-oxidizing phototrophic bacteria. Nat Commun. 2014;5:3391.
Ross DE, Marshall CW, May HD, Norman RS. Metagenome-assembled genome sequences of Acetobacterium sp. strain MES1 and Desulfovibrio sp. strain MES5 from a cathode-associated acetogenic microbial community. Genome Announc. 2017;5:e00938–17.
Patil SA, Arends JBA, Vanwonterghem I, Van Meerbergen J, Guo K, Tyson GW, et al. Selective enrichment establishes a stable performing community for microbial electrosynthesis of acetate from CO2. Environ Sci Technol. 2015;49:8833–43.
Arends JBA, Patil SA, Roume H, Rabaey K. Continuous long-term electricity-driven bioproduction of carboxylates and isopropanol from CO2 with a mixed microbial community. J CO2 Util. 2017;20:141–9.
LaBelle EV, May HD. Energy efficiency and productivity enhancement of microbial electrosynthesis of acetate. Front Microbiol. 2017;8:756.
Arunasri K, Annie Modestra J, Yeruva DK, Vamshi Krishna K, Venkata Mohan S. Polarized potential and electrode materials implication on electro-fermentative di-hydrogen production: microbial assemblages and hydrogenase gene copy variation. Bioresour Technol. 2016;200:691–8.
Molenaar SD, Saha P, Mol AR, Sleutels THJA, ter Heijne A, Buisman CJN. Competition between methanogens and acetogens in biocathodes: a comparison between potentiostatic and galvanostatic control. Int J Mol Sci. 2017;18:pii: E204.
Aryal N, Tremblay P-L, Lizak DM, Zhang T. Performance of different Sporomusa species for the microbial electrosynthesis of acetate from carbon dioxide. Bioresour Technol. 2017;233:184–90.
Rosenbaum MA, Berger C, Schmitz S, Uhlig R. Microbial electrosynthesis i: pure and defined mixed culture engineering. Advances in Biochemical Engineering/Biotechnology. Berlin, Heidelberg: Springer; 2019. pp. 181–202.
Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–80.
Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hügler M, et al. Autotrophic carbon fixation in archaea. Nat Rev Microbiol. 2010;8:447–60.
Fuchs G. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu Rev Microbiol. 2011;65:631–58.
Dietrich K, Dumont M-J, Del Rio LF, Orsat V. Producing PHAs in the bioeconomy—towards a sustainable bioplastic. Sustain Prod Consum. 2017;9:58–70.
Luengo JM, Garcı́a B, Sandoval A, Naharro G, Olivera ER. Bioplastics from microorganisms. Curr Opin Microbiol. 2003;6:251–60.
Możejko-Ciesielska J, Kiewisz R. Bacterial polyhydroxyalkanoates: still fabulous? Microbiol Res. 2016;192:271–82.
Cruz JV, França Z. Hydrogeochemistry of thermal and mineral water springs of the Azores archipelago (Portugal). J Volcanol Geotherm Res. 2006;151:382–98.
Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7:e7359.
Rowe AR, Rajeev P, Jain A, Pirbadian S, Okamoto A, Gralnick JA, et al. Tracking electron uptake from a cathode into Shewanella cells: implications for energy acquisition from solid-substrate electron donors. MBio. 2018;9:e02203–17.
Ross DE, Flynn JM, Baron DB, Gralnick JA, Bond DR. Towards electrosynthesis in Shewanella: energetics of reversing the Mtr pathway for reductive metabolism. PLoS ONE. 2011;6:e16649.
Soussan L, Riess J, Erable B, Delia ML, Bergel A. Electrochemical reduction of CO2 catalysed by Geobacter sulfurreducens grown on polarized stainless steel cathodes. Electrochem Commun. 2013;28:27–30.
Slobodkina GB, Baslerov RV, Novikov AA, Bonch-Osmolovskaya EA, Slobodkin AI. Thermodesulfitimonas autotrophica gen. Nov., sp. Nov., a thermophilic, obligate sulfite-reducing bacterium isolated from a terrestrial hot spring. Int J Syst Evol Microbiol. 2017;67:301–5.
Reiner JE, Jung T, Lapp CJ, Siedler M, Bunk B, Overmann J, et al. Kyrpidia spormannii sp. nov., a thermophilic, hydrogen-oxidizing, facultative autotroph, isolated from hydrothermal systems at São Miguel Island, and emended description of the genus Kyrpidia. Int J Syst Evol Microbiol. 2018;68:3735–40.
Reiner JE, Lapp CJ, Bunk B, Spröer C, Overmann J, Gescher J. Complete genome sequence of Kyrpidia sp. strain EA-1, a thermophilic knallgas bacterium, isolated from the Azores. Genome Announc. 2018;6:e01505–17.
Friedrich T, Scheide D. The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 2000;479:1–5.
Pereira IAC, Ramos AR, Grein F, Marques MC, da Silva SM, Venceslau SS. A comparative genomic analysis of energy metabolism in sulfate reducing bacteria and archaea. Front Microbiol. 2011;2:69.
Battchikova N, Eisenhut M, Aro EM. Cyanobacterial NDH-1 complexes: novel insights and remaining puzzles. Biochim Biophys Acta. 2011;1807:935–44.
Van Keulen G, Girbal L, Van Den Bergh ERE, Dijkhuizen L, Meijer WG. The LysR-type transcriptional regulator CbbR controlling autotrophic CO2 fixation by Xanthobacter flavus is an NADPH sensor. J Bacteriol. 1998;180:1411–7.
Kracke F, Vassilev I, Krömer JO. Microbial electron transport and energy conservation - The foundation for optimizing bioelectrochemical systems. Front Microbiol. 2015;6:1–18.
Allen JWA, Leach N, Ferguson SJ. The histidine of the c-type cytochrome CXXCH haem-binding motif is essential for haem attachment by the Escherichia coli cytochrome c maturation (Ccm) apparatus. Biochem J. 2005;389:587–92.
Shrestha N, Chilkoor G, Vemuri B, Rathinam N, Sani RK, Gadhamshetty V. Extremophiles for microbial-electrochemistry applications: a critical review. Bioresour Technol. 2018;255:318–30.
Faraghiparapari N, Zengler K. Production of organics from CO2 by microbial electrosynthesis (MES) at high temperature. J Chem Technol Biotechnol. 2017;92:375–81.
Dopson M, Ni G, Sleutels THJA. Possibilities for extremophilic microorganisms in microbial electrochemical systems. FEMS Microbiol Rev. 2016;40:164–81.
Logan BE, Rossi R, Ragab A, Saikaly PE. Electroactive microorganisms in bioelectrochemical systems. Nat Rev Microbiol. 2019;17:307–19.
Yu L, Yuan Y, Tang J, Zhou S. Thermophilic Moorella thermoautotrophica-immobilized cathode enhanced microbial electrosynthesis of acetate and formate from CO2. Bioelectrochemistry. 2017;117:23–28.
Stackebrandt E. The emended family Peptococcaceae and description of the families Desulfitobacteriaceae, Desulfotomaculaceae, and Thermincolaceae. In: The prokaryotes. Berlin, Germany: Springer; 2014. pp. 285–90.
Deutzmann JS, Spormann AM. Enhanced microbial electrosynthesis by using defined co-cultures. ISME J. 2017;11:704–14.
Enning D, Garrelfs J. Corrosion of iron by sulfate-reducing bacteria: new views of an old problem. Appl Environ Microbiol. 2014;80:1226–36.
Kato S. Microbial extracellular electron transfer and its relevance to iron corrosion. Microb Biotechnol. 2016;9:141–8.
Zaybak Z, Logan BE, Pisciotta JM. Electrotrophic activity and electrosynthetic acetate production by Desulfobacterium autotrophicum HRM2. Bioelectrochemistry. 2018;123:150–5.
Agostino V, Rosenbaum MA. Sulfate-reducing electroautotrophs and their applications in bioelectrochemical systems. Front Energy Res. 2018;6:55.
Giddings CGS, Nevin KP, Woodward T, Lovley DR, Butler CS. Simplifying microbial electrosynthesis reactor design. Front Microbiol. 2015;6:468.
Plugge CM, Zhang W, Scholten JCM, Stams AJM. Metabolic flexibility of sulfate-reducing bacteria. Front Microbiol. 2011;2:81.
Marshall CW, Ross DE, Handley KM, Weisenhorn PB, Edirisinghe JN, Henry CS, et al. Metabolic reconstruction and modeling microbial electrosynthesis. Sci Rep. 2017;7:8391.
Freas N, Newton P, Perozich J. Analysis of nucleotide diphosphate sugar dehydrogenases reveals family and group-specific relationships. FEBS Open Bio. 2016;6:77–89.
Dangel AW, Tabita FR. CbbR, the master regulator for microbial carbon dioxide fixation. J Bacteriol. 2015;197:3488–98.
Berrisford JM, Baradaran R, Sazanov LA. Structure of bacterial respiratory complex i. Biochim Biophys Acta. 2016;1857:892–901.
Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta. 2011;1807:1398–413.
Carlson HK, Iavarone AT, Gorur A, Yeo BS, Tran R, Melnyk RA, et al. Surface multiheme c-type cytochromes from Thermincola potens and implications for respiratory metal reduction by Gram-positive bacteria. Proc Natl Acad Sci USA. 2012;109:1702–7.
Kurtz DM. Dioxygen-binding proteins. In: Comprehensive coordination chemistry II. Amsterdam, Netherlands: Pergamon; 2004. pp. 229–60.
Volbeda A, Charon MH, Piras C, Hatchikian EC, Frey M, Fontecilla-Camps JC. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature. 1995;373:580–7.
Carbajosa S, Malki M, Caillard R, Lopez MF, Palomares FJ, Martín-Gago JA, et al. Electrochemical growth of Acidithiobacillus ferrooxidans on a graphite electrode for obtaining a biocathode for direct electrocatalytic reduction of oxygen. Biosens Bioelectron. 2010;26:877–80.
de Campos Rodrigues T, Rosenbaum MA. Microbial electroreduction: screening for new cathodic biocatalysts. ChemElectroChem. 2014;1:1916–22.
Krieg T, Sydow A, Faust S, Huth I, Holtmann D. CO2 to terpenes: autotrophic and electroautotrophic α-humulene production with Cupriavidus necator. Angew Chem Int Ed. 2018;57:1879–82.
Bause S, Decker M, Neubauer P, Vonau W. Optimization of the chemolithoautotrophic biofilm growth of Cupriavidus necator by means of electrochemical hydrogen synthesis. Chem Pap. 2018;72:1205–11.
Sturm-Richter K, Golitsch F, Sturm G, Kipf E, Dittrich A, Beblawy S, et al. Unbalanced fermentation of glycerol in Escherichia coli via heterologous production of an electron transport chain and electrode interaction in microbial electrochemical cells. Bioresour Technol. 2015;186:89–96.
Acknowledgements
This work was supported by a grant of the Bundesministerium für Bildung und Forschung (BMBF), no. 033RC006.
Author information
Authors and Affiliations
Contributions
KG performed initial enrichment experiments. MHu and KG designed the specific databases. AD, KG, and JER analyzed the data. WW measured the 13C-content of biofilm samples. JER isolated distinct species from the initial enrichment and performed chronoamperometric experiments with K. spormannii. CJL conducted the K. tusciae control experiment. MF performed SEM experiments. JER conducted and analyzed the comparative transcriptomic analysis of K. spormannii with the help of CJL and TJ. AH-R and MHa designed the flow-cell setup. Flow-cell experiments were performed by JER and MHa. MW provided OCT technology. The paper was written by JER and JG. All authors discussed the results and commented on the paper. SK, HH, and JG conceived and designed the study with inputs from all authors. JG supervised the project.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Reiner, J.E., Geiger, K., Hackbarth, M. et al. From an extremophilic community to an electroautotrophic production strain: identifying a novel Knallgas bacterium as cathodic biofilm biocatalyst. ISME J 14, 1125–1140 (2020). https://doi.org/10.1038/s41396-020-0595-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-020-0595-5
This article is cited by
-
Electron transfer of extremophiles in bioelectrochemical systems
Extremophiles (2022)
-
Bioplastikproduktion mithilfe eines extremophilen kathodischen Biofilms
BIOspektrum (2020)