Abstract
Key message
Glycine soja germplasm can be used to successfully introduce new alleles with the potential to add valuable new genetic diversity to the current elite soybean gene pool.
Abstract
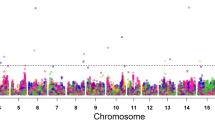
Given the demonstrated narrow genetic base of the US soybean production, it is essential to identify beneficial alleles from exotic germplasm, such as wild soybean, to enhance genetic gain for favorable traits. Nested association mapping (NAM) is an approach to population development that permits the comparison of allelic effects of the same QTL in multiple parents. Seed yield, plant maturity, plant height and plant lodging were evaluated in a NAM panel consisting of 392 recombinant inbred lines derived from three biparental interspecific soybean populations in eight environments during 2016 and 2017. Nested association mapping, combined with linkage mapping, identified three major QTL for plant maturity in chromosomes 6, 11 and 12 associated with alleles from wild soybean resulting in significant increases in days to maturity. A significant QTL for plant height was identified on chromosome 13 with the allele increasing plant height derived from wild soybean. A significant grain yield QTL was detected on chromosome 17, and the allele from Glycine soja had a positive effect of 166 kg ha−1; RIL’s with the wild soybean allele yielded on average 6% more than the lines carrying the Glycine max allele. These findings demonstrate the usefulness and potential of alleles from wild soybean germplasm to enhance important agronomic traits in a soybean breeding program.





Similar content being viewed by others
References
Abdel-Haleem H, Ji P, Boerma HR, Li Z (2013) An R package for SNP marker-based parent-offspring tests. Plant Methods 9:44. https://doi.org/10.1186/1746-4811-9-44
Akpertey A, Belaffif M, Graef GL et al (2014) Effects of selective genetic introgression from wild soybean to soybean. Crop Sci 54:2683–2695. https://doi.org/10.2135/cropsci2014.03.0189
Auer PL, Lettre G (2015) Rare variant association studies: considerations, challenges and opportunities. Genome Med 7:16. https://doi.org/10.1186/s13073-015-0138-2
Bajgain P, Rouse MN, Tsilo TJ et al (2016) Nested association mapping of stem rust resistance in wheat using genotyping by sequencing. PLoS ONE 11:e0155760. https://doi.org/10.1371/journal.pone.0155760
Bandillo NB, Anderson JE, Kantar MB et al (2017) Dissecting the genetic basis of local adaptation in soybean. Sci Rep 7:17195. https://doi.org/10.1038/s41598-017-17342-w
Bernard RL, Cremeens CR (1988) Registration of ‘Williams 82’ soybean. Crop Sci 28:1027–1028. https://doi.org/10.2135/cropsci1988.0011183X002800060049x
Bernardo R (2010) Breeding for quantitative traits in plants, 2nd edn. Stemma Press, Woodbuty
Bradbury PJ, Zhang Z, Kroon DE et al (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Broman KW, Sen S (2009) A guide to QTL mapping with R/qtl. Springer, New York
Buckler ES, Holland JB, Bradbury PJ et al (2009) The genetic architecture of maize flowering time. Science 325(80):714–718. https://doi.org/10.1126/science.1174276
Campos G, Sorensen D, Gianola D (2015) Genomic heritability: what is it? PLoS Genet 11:1–21. https://doi.org/10.1371/journal.pgen.1005048
Cao Y, Li S, Wang Z et al (2017) Identification of major quantitative trait loci for seed oil content in soybeans by combining linkage and genome-wide association mapping. Front Plant Sci 8:1–10. https://doi.org/10.3389/fpls.2017.01222
Carter TE, Hymowitz T, Nelson RL (2004) Biogeography, local adaptation, Vavilov, and genetic diversity in soybean. In: Werner D (ed) Biological resources and migration. Springer, Berlin, pp 47–59
Chen P, Yan K, Shao H, Zhao S (2013) Physiological mechanisms for high salt tolerance in wild soybean (Glycine soja) from Yellow River Delta, China: photosynthesis, osmotic regulation, ion flux and antioxidant capacity. PLoS ONE 8:e83227. https://doi.org/10.1371/journal.pone.0083227
Chen Q, Yang CJ, York AM et al (2019) TeoNAM: a nested association mapping population for domestication and agronomic trait analysis in Maize. bioRxiv 647461. https://doi.org/10.1534/genetics.119.302594
Concibido V, La Vallee B, Mclaird P et al (2003) Introgression of a quantitative trait locus for yield from Glycine soja into commercial soybean cultivars. Theor Appl Genet 106:575–582. https://doi.org/10.1007/s00122-002-1071-5
Diers BW, Specht J, Rainey KM et al (2018) Genetic architecture of soybean yield and agronomic traits. G3 Genes Genomes Genet. https://doi.org/10.1534/g3.118.200332
Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127:1309–1321. https://doi.org/10.1016/j.cell.2006.12.006
Ertl DS, Fehr WR (1985) Agronomic performance of soybean genotypes from Glycine max × Glycine soja Crosses1. Crop Sci 25:589–592. https://doi.org/10.2135/cropsci1985.0011183X002500040003x
Fang C, Ma Y, Wu S et al (2017) Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol 18:161. https://doi.org/10.1186/s13059-017-1289-9
FAOSTAT (2018) Food and Agriculture Organization of the United Nations Statistical Database. http://www.fao.org/faostat/en/#data/QC. Accessed 14 Oct 2018
Fehr WR (1991) Principles of cultivar development: theory and technique, vol 1. Macmillan, New York
Fehr WR, Caviness CE, Burmood DT, Pennington JS (1971) Stage of development descriptions for soybeans, Glycine max (L.) Merrill1. Crop Sci 11:929. https://doi.org/10.2135/cropsci1971.0011183x001100060051x
Flori L, Fritz S, Jaffrézic F et al (2009) The genome response to artificial selection: a case study in dairy cattle. PLoS ONE 4:e6595. https://doi.org/10.1371/journal.pone.0006595
Fragoso CA, Moreno M, Wang Z et al (2017) Genetic architecture of a rice nested association mapping population. G3 Genes Genomes Genet (Bethesda) 7:1913–1926. https://doi.org/10.1534/g3.117.041608
Funatsuki H, Suzuki M, Hirose A et al (2014) Molecular basis of a shattering resistance boosting global dissemination of soybean. Proc Natl Acad Sci USA 111:17797–17802. https://doi.org/10.1073/pnas.1417282111
Gizlice Z, Carter TE, Burton JW (1994) Genetic base for North American Public soybean cultivars released between 1947 and 1988. Crop Sci 34:1143. https://doi.org/10.2135/cropsci1994.0011183X003400050001x
Ha B-K, Kim H-J, Velusamy V et al (2014) Identification of quantitative trait loci controlling linolenic acid concentration in PI483463 (Glycine soja). Theor Appl Genet 127:1501–1512. https://doi.org/10.1007/s00122-014-2314-y
Holland J (2007) Genetic architecture of complex traits in plants. Curr Opin Plant Biol 10:156–161. https://doi.org/10.1016/j.pbi.2007.01.003
Hu Z, Kan G, Zhang G et al (2014) Genetic diversity analysis using simple sequence repeat markers in soybean. Plant Genet Resour 12:S87–S90. https://doi.org/10.1017/S1479262114000331
Hwang E-Y, Song Q, Jia G et al (2014) A genome-wide association study of seed protein and oil content in soybean. BMC Genom 15:1–12. https://doi.org/10.1186/1471-2164-15-1
Hyten DL, Song Q, Zhu Y et al (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci USA 103:16666–16671. https://doi.org/10.1073/pnas.0604379103
Hyten DL, Choi IY, Song Q et al (2007) Highly variable patterns of linkage disequilibrium in multiple soybean populations. Genetics 175:1937–1944. https://doi.org/10.1534/genetics.106.069740
Jarquín D, Kocak K, Posadas L et al (2014) Genotyping by sequencing for genomic prediction in a soybean breeding population. BMC Genom 15:740. https://doi.org/10.1186/1471-2164-15-740
Kang S-T, Kwak M, Kim H-K et al (2009) Population-specific QTLs and their different epistatic interactions for pod dehiscence in soybean [Glycine max (L.) Merr.]. Euphytica 166:15–24. https://doi.org/10.1007/s10681-008-9810-6
Kong F, Liu B, Xia Z et al (2010) Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol 154:1220–1231. https://doi.org/10.1104/pp.110.160796
La T, Large E, Taliercio E et al (2019) Characterization of select wild soybean accessions in the USDA germplasm collection for seed composition and agronomic traits. Crop Sci 59:1–19. https://doi.org/10.2135/cropsci2017.08.0514
Langewisch T, Lenis J, Jiang GL et al (2017) The development and use of a molecular model for soybean maturity groups. BMC Plant Biol 17:91. https://doi.org/10.1186/s12870-017-1040-4
Leamy LJ, Zhang H, Li C et al (2017) A genome-wide association study of seed composition traits in wild soybean (Glycine soja). BMC Genom 18:18. https://doi.org/10.1186/s12864-016-3397-4
Lee SH, Bailey MA, Mian MAR et al (1996) Molecular markers associated with soybean plant height, lodging, and maturity across locations. Crop Sci 36:728–735. https://doi.org/10.2135/cropsci1996.0011183X003600030035x
Lee J-D, Shannon JG, Vuong TD, Nguyen HT (2009) Inheritance of salt tolerance in wild soybean (Glycine soja Sieb. and Zucc.) Accession PI483463. J Hered 100:798–801. https://doi.org/10.1093/jhered/esp027
Li D, Pfeiffer TW, Cornelius PL (2008) Soybean QTL for yield and yield components associated with alleles. Crop Sci 48:571. https://doi.org/10.2135/cropsci2007.06.0361
Li S, Cao Y, He J et al (2017) Detecting the QTL-allele system conferring flowering date in a nested association mapping population of soybean using a novel procedure. Theor Appl Genet 130:2297–2314. https://doi.org/10.1007/s00122-017-2960-y
Li MW, Liu W, Lam HM, Gendron JM (2019) Characterization of two growth period QTLs reveals modification of PRR3 genes during soybean domestication. Plant Cell Physiol 60:407–420. https://doi.org/10.1093/pcp/pcy215
Liu B, Fujita T, Yan Z-H et al (2007) QTL mapping of domestication-related traits in soybean (Glycine max). Ann Bot 100:1027–1038. https://doi.org/10.1093/aob/mcm149
Liu B, Kanazawa A, Matsumura H et al (2008) Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180:995–1007. https://doi.org/10.1534/genetics.108.092742
Mao T, Li J, Wen Z et al (2017) Association mapping of loci controlling genetic and environmental interaction of soybean flowering time under various photo-thermal conditions. BMC Genom 18:415. https://doi.org/10.1186/s12864-017-3778-3
Maurer A, Sannemann W, Léon J, Pillen K (2017) Estimating parent-specific QTL effects through cumulating linked identity-by-state SNP effects in multiparental populations. Heredity (Edinb) 118:477–485. https://doi.org/10.1038/hdy.2016.121
Ning W, Zhai H, Yu J et al (2017) Overexpression of Glycine soja WRKY20 enhances drought tolerance and improves plant yields under drought stress in transgenic soybean. Mol Breed 37:19. https://doi.org/10.1007/s11032-016-0614-4
Nisa Z, Mallano A, Yu Y et al (2017) GsSNAP33, a novel Glycine soja SNAP25-type protein gene: improvement of plant salt and drought tolerances in transgenic Arabidopsis thaliana. Plant Physiol Biochem 119:9–20. https://doi.org/10.1016/j.plaphy.2017.07.029
Price AL, Zaitlen NA, Reich D, Patterson N (2010) New approaches to population stratification in genome-wide association studies. Nat Rev Genet 11:459–463. https://doi.org/10.1038/nrg2813
Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8:e66428. https://doi.org/10.1371/journal.pone.0066428
Reinprecht Y, Poysa VW, Yu K et al (2006) Seed and agronomic QTL in low linolenic acid, lipoxygenase-free soybean (Glycine max (L.) Merrill) germplasm. Genome 49:1510–1527. https://doi.org/10.1139/g06-112
Rincker K, Nelson R, Specht J et al (2014) Genetic improvement of U.S. soybean in maturity groups II, III, and IV. Crop Sci 54:1419–1432. https://doi.org/10.2135/cropsci2013.10.0665
Rogers J, Chen P, Shi A et al (2015) Agronomic performance and genetic progress of selected historical soybean varieties in the southern USA. Plant Breed 134:85–93. https://doi.org/10.1111/pbr.12222
Rowntree SC, Suhre JJ, Weidenbenner NH et al (2013) Genetic gain × management interactions in soybean: I. Planting date. Crop Sci 53:1128. https://doi.org/10.2135/cropsci2012.03.0157
Sharma R, Draicchio F, Bull H et al (2018) Genome-wide association of yield traits in a nested association mapping population of barley reveals new gene diversity for future breeding. J Exp Bot 69:3811–3822. https://doi.org/10.1093/jxb/ery178
Song Q, Hyten DL, Jia G et al (2013) Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS ONE 8:e54985. https://doi.org/10.1371/journal.pone.0054985
Song Q, Hyten DL, Jia G et al (2015) Fingerprinting soybean germplasm and its utility in genomic research. G3 Genes Genomes Genet (Bethesda) 5:1999–2006. https://doi.org/10.1534/g3.115.019000
Song Q, Jenkins J, Jia G et al (2016) Construction of high resolution genetic linkage maps to improve the soybean genome sequence assembly Glyma1.01. BMC Genomics 17:33. https://doi.org/10.1186/s12864-015-2344-0
Specht JE, Chase K, Macrander M et al (2001) Soybean response to water: a QTL analysis of drought tolerance. Crop Sci 41:493–509. https://doi.org/10.2135/cropsci2001.412493x
Tan B, Grattapaglia D, Martins GS et al (2017) Evaluating the accuracy of genomic prediction of growth and wood traits in two Eucalyptus species and their F1 hybrids. BMC Plant Biol. https://doi.org/10.1186/s12870-017-1059-6
United Department of Agriculture (USDA) (2018) National agriculture statistics. https://quickstats.nass.usda.gov/results. Accessed 23 Sept 2018
Vatter T, Maurer A, Kopahnke D et al (2017) A nested association mapping population identifies multiple small effect QTL conferring resistance against net blotch (Pyrenophora teres f. teres) in wild barley. PLoS ONE. https://doi.org/10.1371/journal.pone.0186803
Wang D, Diers BW, Arelli PR, Shoemaker RC (2001) Loci underlying resistance to Race 3 of soybean cyst nematode in Glycine soja plant introduction 468916. Theor Appl Genet 103:561–566. https://doi.org/10.1007/PL00002910
Wang D, Procopiuk AM, Diers BW, Graef GL (2004) Identification of putative QTL that underlie yield in interspecific soybean backcross populations. TAG Theor Appl Genet 108:458–467. https://doi.org/10.1007/s00122-003-1449-z
Wang X, Jiang G-L, Green M et al (2014) Identification and validation of quantitative trait loci for seed yield, oil and protein contents in two recombinant inbred line populations of soybean. Mol Genet Genomics 289:935–949. https://doi.org/10.1007/s00438-014-0865-x
Wei J, Xu S (2016) A random-model approach to QTL mapping in multiparent advanced generation intercross (MAGIC) populations. Genetics 202:471–486. https://doi.org/10.1534/genetics.115.179945
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution (N Y) 38:1358. https://doi.org/10.2307/2408641
Wen Z, Ding Y, Zhao T-J, Gai J (2009) Genetic diversity and peculiarity of annual wild soybean (G. soja Sieb. et Zucc.) from various eco-regions in China. Theor Appl Genet 119:371–381. https://doi.org/10.1007/s00122-009-1045-y
Winter SMJ, Shelp BJ, Anderson TR et al (2007) QTL associated with horizontal resistance to soybean cyst nematode in Glycine soja PI464925B. Theor Appl Genet 114:461–472. https://doi.org/10.1007/s00122-006-0446-4
Wolfgang G, An YC (2017) Genetic separation of southern and northern soybean breeding programs in North America and their associated allelic variation at four maturity loci. Mol Breed 37:8. https://doi.org/10.1007/s11032-016-0611-7
Xavier A, Xu S, Muir WM, Rainey KM (2015) NAM: association studies in multiple populations. Bioinformatics 31:btv448. https://doi.org/10.1093/bioinformatics/btv448
Xavier A, Hall B, Hearst AA et al (2017) Genetic architecture of phenomic-enabled canopy coverage in Glycine max. Genetics 206:1081–1089. https://doi.org/10.1534/genetics.116.198713
Xavier A, Jarquin D, Howard R et al (2018) Genome-wide analysis of grain yield stability and environmental interactions in a multiparental soybean population. G3 Genes Genomes Genet (Bethesda) 8:519–529. https://doi.org/10.1534/g3.117.300300
Xia Z, Watanabe S, Yamada T et al (2012) Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc Natl Acad Sci 109:E2155–E2164. https://doi.org/10.1073/pnas.1117982109
Xu M, Xu Z, Liu B et al (2013) Genetic variation in four maturity genes affects photoperiod insensitivity and PHYA-regulated post-flowering responses of soybean. BMC Plant Biol 13:91. https://doi.org/10.1186/1471-2229-13-91
Xu Y, Li P, Yang Z, Xu C (2017) Genetic mapping of quantitative trait loci in crops. Crop J 5:175–184. https://doi.org/10.1016/j.cj.2016.06.003
Yan L, Xing L-L, Yang C-Y et al (2014) Identification of quantitative trait loci associated with soybean seed protein content using two populations derived from crosses between Glycine max and Glycine soja. Plant Genet Resour 12:S104–S108. https://doi.org/10.1017/S1479262114000379
Yan L, Hofmann N, Li S et al (2017) Identification of QTL with large effect on seed weight in a selective population of soybean with genome-wide association and fixation index analyses. BMC Genom 18:1–11. https://doi.org/10.1186/s12864-017-3922-0
Yu J, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178:539–551. https://doi.org/10.1534/genetics.107.074245
Zhang J, Song Q, Cregan PB et al (2015) Genome-wide association study for flowering time, maturity dates and plant height in early maturing soybean (Glycine max) germplasm. BMC Genom 16:217. https://doi.org/10.1186/s12864-015-1441-4
Zhang H, Li C, Davis EL et al (2016) Genome-wide association study of resistance to soybean cyst nematode (Heterodera glycines) HG type 2.5.7 in wild soybean (Glycine soja). Front Plant Sci 7:1–11. https://doi.org/10.3389/fpls.2016.01214
Zhang H, Mittal N, Leamy LJ et al (2017a) Back into the wild—apply untapped genetic diversity of wild relatives for crop improvement. Evol Appl 10:5–24
Zhang H, Song Q, Griffin JD, Song B-H (2017b) Genetic architecture of wild soybean (Glycine soja) response to soybean cyst nematode (Heterodera glycines). Mol Genet Genomics 292:1257–1265. https://doi.org/10.1007/s00438-017-1345-x
Zhang S, Zhang Z, Bales C et al (2017c) Mapping novel aphid resistance QTL from wild soybean, Glycine soja 85-32. Theor Appl Genet 130:1941–1952. https://doi.org/10.1007/s00122-017-2935-z
Zhang X, Wang W, Guo N et al (2018) Combining QTL-seq and linkage mapping to fine map a wild soybean allele characteristic of greater plant height. BMC Genom 19:226. https://doi.org/10.1186/s12864-018-4582-4
Zhou Z, Jiang Y, Wang Z et al (2015) Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol 33:408–414. https://doi.org/10.1038/nbt.3096
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome J 1:5. https://doi.org/10.3835/plantgenome2008.02.0089
Acknowledgements
The authors would like to acknowledge funding from the Missouri Soybean Merchandising Council and the United Soybean Board. We also thank personnel from the soybean breeding program at the University of Missouri for their time and effort in preparing and conducting the experiments in 2016 and 2017.
Author information
Authors and Affiliations
Contributions
EB conducted field evaluations and data analysis; AMS acquired funding and supervised the work; QS performed the genotyping; RN developed the initial populations; GS revised the manuscript; JDG, TB, and JD provided statistical expertise and revised the manuscript; EB, AMS, and JG wrote the paper. All authors read the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Albrecht E. Melchinger.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Randall Nelson was retired from USDA-Agricultural Research Service.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Beche, E., Gillman, J.D., Song, Q. et al. Nested association mapping of important agronomic traits in three interspecific soybean populations. Theor Appl Genet 133, 1039–1054 (2020). https://doi.org/10.1007/s00122-019-03529-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03529-4