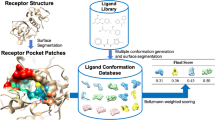
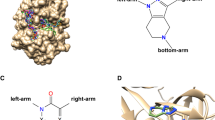
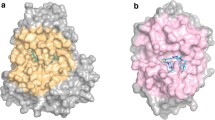
Abstract
The Drug Design Data Resource (D3R) aims to identify best practice methods for computer aided drug design through blinded ligand pose prediction and affinity challenges. Herein, we report on the results of Grand Challenge 4 (GC4). GC4 focused on proteins beta secretase 1 and Cathepsin S, and was run in an analogous manner to prior challenges. In Stage 1, participant ability to predict the pose and affinity of BACE1 ligands were assessed. Following the completion of Stage 1, all BACE1 co-crystal structures were released, and Stage 2 tested affinity rankings with co-crystal structures. We provide an analysis of the results and discuss insights into determined best practice methods.









Similar content being viewed by others
References
Macalino SJY, Gosu V, Hong S, Choi S (2015) Role of computer-aided drug design in modern drug discovery. Arch Pharm Res 38:1686–1701. https://doi.org/10.1007/s12272-015-0640-5
Jorgensen WL (2004) The many roles of computation in drug discovery. Science 303:1813–1818. https://doi.org/10.1126/science.1096361
Sliwoski G, Kothiwale S, Meiler J, Lowe EW (2013) Computational methods in drug discovery. Pharmacol Rev 66:334–395. https://doi.org/10.1124/pr.112.007336
Irwin JJ, Shoichet BK (2016) Docking screens for novel ligands conferring new biology. J Med Chem 59:4103–4120. https://doi.org/10.1021/acs.jmedchem.5b02008
Liao C, Sitzmann M, Pugliese A, Nicklaus MC (2011) Software and resources for computational medicinal chemistry. Fut Med Chem 3:1057–1085. https://doi.org/10.4155/fmc.11.63
Gaieb Z, Liu S, Gathiaka S et al (2018) D3R Grand Challenge 2: blind prediction of protein–ligand poses, affinity rankings, and relative binding free energies. J Comput Aided Mol Des 32:1–20. https://doi.org/10.1007/s10822-017-0088-4
Athanasiou C, Vasilakaki S, Dellis D, Cournia Z (2018) Using physics-based pose predictions and free energy perturbation calculations to predict binding poses and relative binding affinities for FXR ligands in the D3R Grand Challenge 2. J Comput Aided Mol Des 32:21–44. https://doi.org/10.1007/s10822-017-0075-9
Baumgartner MP, Evans DA (2018) Lessons learned in induced fit docking and metadynamics in the Drug Design Data Resource Grand Challenge 2. J Comput Aided Mol Des 32:45–58. https://doi.org/10.1007/s10822-017-0081-y
Bhakat S, Åberg E, Söderhjelm P (2018) Prediction of binding poses to FXR using multi-targeted docking combined with molecular dynamics and enhanced sampling. J Comput Aided Mol Des 32:59–73. https://doi.org/10.1007/s10822-017-0074-x
da Silva Figueiredo Celestino Gomes P, Da Silva F, Bret G, Rognan D (2018) Ranking docking poses by graph matching of protein–ligand interactions: lessons learned from the D3R Grand Challenge 2. J Comput Aided Mol Des 32:75–87. https://doi.org/10.1007/s10822-017-0046-1
Ding X, Hayes RL, Vilseck JZ et al (2018) CDOCKER and $$\lambda$$λ-dynamics for prospective prediction in D3R Grand Challenge 2. J Comput Aided Mol Des 32:89–102. https://doi.org/10.1007/s10822-017-0050-5
Duan R, Xu X, Zou X (2018) Lessons learned from participating in D3R 2016 Grand Challenge 2: compounds targeting the farnesoid X receptor. J Comput Aided Mol Des 32:103–111. https://doi.org/10.1007/s10822-017-0082-x
Fradera X, Verras A, Hu Y et al (2018) Performance of multiple docking and refinement methods in the pose prediction D3R prospective Grand Challenge 2016. J Comput Aided Mol Des 32:113–127. https://doi.org/10.1007/s10822-017-0053-2
Gao Y-D, Hu Y, Crespo A et al (2018) Workflows and performances in the ranking prediction of 2016 D3R Grand Challenge 2: lessons learned from a collaborative effort. J Comput Aided Mol Des 32:129–142. https://doi.org/10.1007/s10822-017-0072-z
Hogues H, Sulea T, Gaudreault F et al (2018) Binding pose and affinity prediction in the 2016 D3R Grand Challenge 2 using the Wilma-SIE method. J Comput Aided Mol Des 32:143–150. https://doi.org/10.1007/s10822-017-0071-0
Kadukova M, Grudinin S (2018) Docking of small molecules to farnesoid X receptors using AutoDock Vina with the Convex-PL potential: lessons learned from D3R Grand Challenge 2. J Comput Aided Mol Des 32:151–162. https://doi.org/10.1007/s10822-017-0062-1
Kumar A, Zhang KYJ (2018) A cross docking pipeline for improving pose prediction and virtual screening performance. J Comput Aided Mol Des 32:163–173. https://doi.org/10.1007/s10822-017-0048-z
Kurkcuoglu Z, Koukos PI, Citro N et al (2018) Performance of HADDOCK and a simple contact-based protein–ligand binding affinity predictor in the D3R Grand Challenge 2. J Comput Aided Mol Des 32:175–185. https://doi.org/10.1007/s10822-017-0049-y
Lam PC-H, Abagyan R, Totrov M (2018) Ligand-biased ensemble receptor docking (LigBEnD): a hybrid ligand/receptor structure-based approach. J Comput Aided Mol Des 32:187–198. https://doi.org/10.1007/s10822-017-0058-x
Mey ASJS, Jiménez JJ, Michel J (2018) Impact of domain knowledge on blinded predictions of binding energies by alchemical free energy calculations. J Comput Aided Mol Des 32:199–210. https://doi.org/10.1007/s10822-017-0083-9
Olsson MA, García-Sosa AT, Ryde U (2018) Binding affinities of the farnesoid X receptor in the D3R Grand Challenge 2 estimated by free-energy perturbation and docking. J Comput Aided Mol Des 32:211–224. https://doi.org/10.1007/s10822-017-0056-z
Padhorny D, Hall DR, Mirzaei H et al (2018) Protein–ligand docking using FFT based sampling: D3R case study. J Comput Aided Mol Des 32:225–230. https://doi.org/10.1007/s10822-017-0069-7
Réau M, Langenfeld F, Zagury J-F, Montes M (2018) Predicting the affinity of Farnesoid X receptor ligands through a hierarchical ranking protocol: a D3R Grand Challenge 2 case study. J Comput Aided Mol Des 32:231–238. https://doi.org/10.1007/s10822-017-0063-0
Rifai EA, van Dijk M, Vermeulen NPE, Geerke DP (2018) Binding free energy predictions of farnesoid X receptor (FXR) agonists using a linear interaction energy (LIE) approach with reliability estimation: application to the D3R Grand Challenge 2. J Comput Aided Mol Des 32:239–249. https://doi.org/10.1007/s10822-017-0055-0
Salmaso V, Sturlese M, Cuzzolin A, Moro S (2018) Combining self- and cross-docking as benchmark tools: the performance of DockBench in the D3R Grand Challenge 2. J Comput Aided Mol Des 32:251–264. https://doi.org/10.1007/s10822-017-0051-4
Schindler C, Rippmann F, Kuhn D (2018) Relative binding affinity prediction of farnesoid X receptor in the D3R Grand Challenge 2 using FEP+. J Comput Aided Mol Des 32:265–272. https://doi.org/10.1007/s10822-017-0064-z
Selwa E, Elisée E, Zavala A, Iorga BI (2018) Blinded evaluation of farnesoid X receptor (FXR) ligands binding using molecular docking and free energy calculations. J Comput Aided Mol Des 32:273–286. https://doi.org/10.1007/s10822-017-0054-1
Wingert BM, Oerlemans R, Camacho CJ (2018) Optimal affinity ranking for automated virtual screening validated in prospective D3R grand challenges. J Comput Aided Mol Des 32:287–297. https://doi.org/10.1007/s10822-017-0065-y
Yakovenko O, Jones SJM (2017) Modern drug design: the implication of using artificial neuronal networks and multiple molecular dynamic simulations. J Comput Aided Mol Des 32:299–311. https://doi.org/10.1007/s10822-017-0085-7
Gaieb Z, Parks CD, Chiu M et al (2019) D3R Grand Challenge 3: blind prediction of protein–ligand poses and affinity rankings. J Comput Aided Mol Des. https://doi.org/10.1007/s10822-018-0180-4
Sunseri J, King JE, Francoeur PG, Koes DR (2019) Convolutional neural network scoring and minimization in the D3R 2017 community challenge. J Comput Aided Mol Des 33:19–34. https://doi.org/10.1007/s10822-018-0133-y
Lam PC-H, Abagyan R, Totrov M (2019) Hybrid receptor structure/ligand-based docking and activity prediction in ICM: development and evaluation in D3R Grand Challenge 3. J Comput Aided Mol Des 33:35–46. https://doi.org/10.1007/s10822-018-0139-5
Kumar A, Zhang KYJ (2019) Shape similarity guided pose prediction: lessons from D3R Grand Challenge 3. J Comput Aided Mol Des 33:47–59. https://doi.org/10.1007/s10822-018-0142-x
Xie B, Minh DDL (2019) Alchemical Grid Dock (AlGDock) calculations in the D3R Grand Challenge 3. J Comput Aided Mol Des 33:61–69. https://doi.org/10.1007/s10822-018-0143-9
Nguyen DD, Cang Z, Wu K et al (2019) Mathematical deep learning for pose and binding affinity prediction and ranking in D3R Grand Challenges. J Comput Aided Mol Des 33:71–82. https://doi.org/10.1007/s10822-018-0146-6
Koukos PI, Xue LC, Bonvin AMJJ (2019) Protein–ligand pose and affinity prediction: lessons from D3R Grand Challenge 3. J Comput Aided Mol Des 33:83–91. https://doi.org/10.1007/s10822-018-0148-4
Chaput L, Selwa E, Elisée E, Iorga BI (2019) Blinded evaluation of cathepsin S inhibitors from the D3RGC3 dataset using molecular docking and free energy calculations. J Comput Aided Mol Des 33:93–103. https://doi.org/10.1007/s10822-018-0161-7
He X, Man VH, Ji B et al (2019) Calculate protein–ligand binding affinities with the extended linear interaction energy method: application on the Cathepsin S set in the D3R Grand Challenge 3. J Comput Aided Mol Des 33:105–117. https://doi.org/10.1007/s10822-018-0162-6
Ignatov M, Liu C, Alekseenko A et al (2019) Monte Carlo on the manifold and MD refinement for binding pose prediction of protein–ligand complexes: 2017 D3R Grand Challenge. J Comput Aided Mol Des 33:119–127. https://doi.org/10.1007/s10822-018-0176-0
Wallach I, Heifets A (2018) Most ligand-based classification benchmarks reward memorization rather than generalization. J Chem Inf Model 58:916–932. https://doi.org/10.1021/acs.jcim.7b00403
Carlson HA (2016) Lessons learned over four benchmark exercises from the community structure–activity resource. J Chem Inf Model 56:951–954. https://doi.org/10.1021/acs.jcim.6b00182
Carlson HA, Smith RD, Damm-Ganamet KL et al (2016) CSAR 2014: a benchmark exercise using unpublished data from pharma. J Chem Inf Model 56:1063–1077. https://doi.org/10.1021/acs.jcim.5b00523
Damm-Ganamet KL, Smith RD, Dunbar JB et al (2013) CSAR benchmark exercise 2011–2012: evaluation of results from docking and relative ranking of blinded congeneric series. J Chem Inf Model 53:1853–1870. https://doi.org/10.1021/ci400025f
Smith RD, Damm-Ganamet KL, Dunbar JB et al (2016) CSAR benchmark exercise 2013: evaluation of results from a combined computational protein design, docking, and scoring/ranking challenge. J Chem Inf Model 56:1022–1031. https://doi.org/10.1021/acs.jcim.5b00387
Gathiaka S, Liu S, Chiu M et al (2016) D3R grand challenge 2015: evaluation of protein–ligand pose and affinity predictions. J Comput Aided Mol Des 30:651–668. https://doi.org/10.1007/s10822-016-9946-8
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23:3–25. https://doi.org/10.1016/S0169-409X(96)00423-1
Veber DF, Johnson SR, Cheng H-Y et al (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45:2615–2623. https://doi.org/10.1021/jm020017n
RDKit (2019) https://www.rdkit.org/. Accessed 13 Aug 2019
Yu HS, Deng Y, Wu Y et al (2017) Accurate and reliable prediction of the binding affinities of macrocycles to their protein targets. J Chem Theory Comput 13:6290–6300. https://doi.org/10.1021/acs.jctc.7b00885
Machauer R, Laumen K, Veenstra S et al (2009) Macrocyclic peptidomimetic β-secretase (BACE-1) inhibitors with activity in vivo. Bioorg Med Chem Lett 19:1366–1370. https://doi.org/10.1016/j.bmcl.2009.01.055
Hanessian S, Yang G, Rondeau J-M et al (2006) Structure-based design and synthesis of macroheterocyclic peptidomimetic inhibitors of the aspartic protease β-site amyloid precursor protein cleaving enzyme (BACE). J Med Chem 49:4544–4567. https://doi.org/10.1021/jm060154a
Thurmond RL, Sun S, Sehon CA et al (2004) Identification of a potent and selective noncovalent cathepsin S inhibitor. J Pharmacol Exp Ther 308:268–276. https://doi.org/10.1124/jpet.103.056879
Jacobson MP, Pincus DL, Rapp CS, Day TJ, Honig B, Shaw DE, Friesner RA (2004) A hierarchical approach to all-atom protein loop prediction. Proteins 55:351–367. https://doi.org/10.1002/prot.10613
Kendall MG (1938) A new measure of rank correlation. Biometrika 30:81–93. https://doi.org/10.2307/2332226
Kendall MG (1945) The treatment of ties in ranking problems. Biometrika 33:239–251. https://doi.org/10.2307/2332303
Zwillinger D, Kokoska S (1999) CRC standard probability and statistics tables and formulae. CRC Press, Boca Raton
Wildman SA, Crippen GM (1999) Prediction of physicochemical parameters by atomic contributions. J Chem Inf Comput Sci 39:868–873. https://doi.org/10.1021/ci990307l
Breiman L (2001) Random forests. Mach Learn 45:5–32. https://doi.org/10.1023/A:1010933404324
Pedregosa F, Varoquaux G, Gramfort A et al (2011) Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830
Buitinck L, Louppe G, Blondel M et al (2013) API design for machine learning software: experiences from the scikit-learn project. arXiv:13090238
Gaulton A, Hersey A, Nowotka M et al (2017) The ChEMBL database in 2017. Nucleic Acids Res 45:D945–D954. https://doi.org/10.1093/nar/gkw1074
Das B, Yan R (2017) Role of BACE1 in Alzheimer’s synaptic function. Transl Neurodegener 6:23. https://doi.org/10.1186/s40035-017-0093-5
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334
Friesner RA, Banks JL, Murphy RB et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749. https://doi.org/10.1021/jm0306430
Halgren TA, Murphy RB, Friesner RA et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem 47:1750–1759. https://doi.org/10.1021/jm030644s
Abagyan R, Totrov M, Kuznetsov D (1994) ICM—a new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J Comput Chem 15:488–506. https://doi.org/10.1002/jcc.540150503
Tetko IV, Gasteiger J, Todeschini R et al (2005) Virtual computational chemistry laboratory—design and description. J Comput Aided Mol Des 19:453–463. https://doi.org/10.1007/s10822-005-8694-y
Verdonk ML, Cole JC, Hartshorn MJ et al (2003) Improved protein–ligand docking using GOLD. Proteins 52:609–623. https://doi.org/10.1002/prot.10465
Ihlenfeldt WD, Takahashi Y, Abe H, Sasaki SI (1994) Computation and management of chemical properties in CACTVS: an extensible networked approach toward modularity and compatibility. J Chem Inf Model 34:109–116. https://doi.org/10.1021/ci00017a013
Chaudhury S, Gray JJ (2008) Conformer selection and induced fit in flexible backbone protein-protein docking using computational and NMR ensembles. J Mol Biol 381:1068–1087. https://doi.org/10.1016/j.jmb.2008.05.042
Feinstein WP, Brylinski M (2014) e FindSite: enhanced fingerprint-based virtual screening against predicted ligand binding sites in protein models. Mol Inform 33:135–150. https://doi.org/10.1002/minf.201300143
Coutsias EA, Lexa KW, Wester MJ et al (2016) Exhaustive conformational sampling of complex fused ring macrocycles using inverse kinematics. J Chem Theory Comput 12:4674–4687. https://doi.org/10.1021/acs.jctc.6b00250
O’Boyle NM, Banck M, James CA et al (2011) Open babel: an open chemical toolbox. J Cheminform 3:33. https://doi.org/10.1186/1758-2946-3-33
Maestro, Schrödinger Release 2019-4. Schrödinger, New York
Bonnet P, Agrafiotis DK, Zhu F, Martin E (2009) Conformational analysis of macrocycles: finding what common search methods miss. J Chem Inf Model 49:2242–2259. https://doi.org/10.1021/ci900238a
Morris GM, Huey R, Lindstrom W et al (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. https://doi.org/10.1002/jcc.21256
Amaro RE, Baudry J, Chodera J et al (2018) Ensemble docking in drug discovery. Biophys J 114:2271–2278. https://doi.org/10.1016/j.bpj.2018.02.038
Amaro RE, Baron R, McCammon JA (2008) An improved relaxed complex scheme for receptor flexibility in computer-aided drug design. J Comput Aided Mol Des 22:693–705. https://doi.org/10.1007/s10822-007-9159-2
Korb O, Olsson TSG, Bowden SJ et al (2012) Potential and limitations of ensemble docking. J Chem Inf Model 52:1262–1274. https://doi.org/10.1021/ci2005934
Tuccinardi T, Botta M, Giordano A, Martinelli A (2010) Protein kinases: docking and homology modeling reliability. J Chem Inf Model 50:1432–1441. https://doi.org/10.1021/ci100161z
Jones G, Willett P, Glen RC et al (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267:727–748. https://doi.org/10.1006/jmbi.1996.0897
Jiménez J, Škalič M, Martínez-Rosell G, De Fabritiis G (2018) KDEEP: protein–ligand absolute binding affinity prediction via 3D-convolutional neural networks. J Chem Inf Model 58:287–296. https://doi.org/10.1021/acs.jcim.7b00650
Abagyan R, Totrov M (1994) Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol 235:983–1002. https://doi.org/10.1006/jmbi.1994.1052
Totrov M, Abagyan R (1999) Derivation of sensitive discrimination potential for virtual ligand screening. In: Proceedings of the third annual international conference on Computational molecular biology—RECOMB ’99. ACM Press, Lyon, pp 312–320
Bramer D, Wei G-W (2018) Multiscale weighted colored graphs for protein flexibility and rigidity analysis. J Chem Phys 148:054103. https://doi.org/10.1063/1.5016562
Nguyen DD, Xiao T, Wang M, Wei G-W (2017) Rigidity strengthening: a mechanism for protein–ligand binding. J Chem Inf Model 57:1715–1721. https://doi.org/10.1021/acs.jcim.7b00226
Cang Z, Wei G-W (2018) Integration of element specific persistent homology and machine learning for protein-ligand binding affinity prediction. Int J Numer Methods Biomed Eng 34:e2914. https://doi.org/10.1002/cnm.2914
Cang Z, Wei G-W (2017) TopologyNet: topology based deep convolutional and multi-task neural networks for biomolecular property predictions. PLoS Comput Biol 13:e1005690. https://doi.org/10.1371/journal.pcbi.1005690
Cang Z, Mu L, Wei G-W (2018) Representability of algebraic topology for biomolecules in machine learning based scoring and virtual screening. PLoS Comput Biol 14:e1005929. https://doi.org/10.1371/journal.pcbi.1005929
Jorgensen WL, Thomas LL (2008) Perspective on free-energy perturbation calculations for chemical equilibria. J Chem Theory Comput 4:869–876. https://doi.org/10.1021/ct800011m
Mobley DL, Klimovich PV (2012) Perspective: alchemical free energy calculations for drug discovery. J Chem Phys 137:230901. https://doi.org/10.1063/1.4769292
Chodera JD, Mobley DL, Shirts MR et al (2011) Alchemical free energy methods for drug discovery: progress and challenges. Curr Opin Struct Biol 21:150–160. https://doi.org/10.1016/j.sbi.2011.01.011
Cournia Z, Allen B, Sherman W (2017) Relative binding free energy calculations in drug discovery: recent advances and practical considerations. J Chem Inf Model 57:2911–2937. https://doi.org/10.1021/acs.jcim.7b00564
Christ CD, Fox T (2014) Accuracy assessment and automation of free energy calculations for drug design. J Chem Inf Model 54:108–120. https://doi.org/10.1021/ci4004199
Mobley DL, Gilson MK (2017) Predicting binding free energies: frontiers and benchmarks. Annu Rev Biophys 46:531–558. https://doi.org/10.1146/annurev-biophys-070816-033654
Acknowledgements
This work was supported by National Institutes of Health (NIH) Grant 1U01GM111528 for the Drug Design Data Resource (D3R). We also thank OpenEye Scientific Software for generously donating the use of their software. We thank Prof. William Jorgensen (Yale) for providing valuable insight into the selected free energy sets. The RCSB PDB is jointly funded by the National Science Foundation (DBI-1832184), the National Institutes of Health (R01GM133198), and the United States Department of Energy (DE-SC0019749).The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the federal funding agencies. MKG has an equity interest in, and is a co-founder and scientific advisor of, VeraChem LLC; REA has equity interest in and is a co-founder and scientific advisor of Actavalon, Inc.; and PW has an equity interest in Relay Pharmaceuticals, Inc. We also thank the reviewers for their helpful suggestions.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Parks, C.D., Gaieb, Z., Chiu, M. et al. D3R grand challenge 4: blind prediction of protein–ligand poses, affinity rankings, and relative binding free energies. J Comput Aided Mol Des 34, 99–119 (2020). https://doi.org/10.1007/s10822-020-00289-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-020-00289-y