Abstract
Obesity, caused by the dysfunction of white adipose tissue (WAT), is reportedly accompanied by exacerbation of lipolysis. Perilipin 1 (PLIN1), which forms a coat around lipid droplets, interacts with several lipolysis proteins to regulate lipolysis. While it is known that perilipin family proteins are degraded in lysosomes, the underlying molecular mechanisms related to the downregulated expression of PLIN1 in obese WAT remain unknown. Recently, we found that lysosomal dysfunction originating from an abnormality of cathepsin B (CTSB), a lysosomal representative protease, occurs in obese WAT. Therefore, we investigated the effect of CTSB alterations on PLIN1 expression in obese WAT. PLIN1 protein disappeared and CTSB protein appeared in the cytoplasm of adipocytes in the early stage of obese WAT. Overexpression of CTSB reduced PLIN1 protein in 3T3L1 adipocytes, and treatment with a CTSB inhibitor significantly recovered this reduction. In addition, CTSB overexpression induced the dysfunction of lipolysis in 3T3L1 adipocytes. Therefore, we concluded that upregulation of CTSB induced the reduction of PLIN1 protein in obese WAT, resulting in lipolysis dysfunction. This suggests a novel pathology of lipid metabolism involving PLIN1 in adipocytes and that CTSB might be a therapeutic candidate molecule for obese WAT.
Similar content being viewed by others
Introduction
White adipose tissue (WAT) is composed of many adipocytes and functions to store surplus fuel as lipid droplets (LDs) and supply it as required, but excess WAT leads to obesity1,2,3. Perilipin coats LDs in adipocytes, thus playing a role in their stabilization and lipolysis by lipases and their cofactors4. The perilipin family is composed of five proteins: perilipin 1 (PLIN1), perilipin 2 (adipophilin/PLIN2), perilipin 3 (TIP47/PLIN3), perilipin 4 (S3-12/PLIN4), and perilipin 5 (OXPAT/MLDP/PLIN5). PLIN1 is highly expressed in WAT and is the most studied member of the perilipin family4,5,6. Under lipolytic conditions such as isoproterenol treatment, cAMP-dependent protein kinase phosphorylates hormone-sensitive lipase (HSL) and PLIN1. Phosphorylation of HSL induces its catabolic activity and translocation from the cytosol to LD, while phosphorylation of PLIN1 changes its conformation to facilitate the attachment of activated HSL to LD, leading to the hydrolysis of triacylglycerol to diacylglycerol and fatty acid7,8,9. Notably, the phosphorylation of PLIN1 at serine (Ser)-492 and Ser-517 was associated with basal and hormone-stimulated lipolysis7,8,9. Moreover, mice with a systemic deletion of PLIN1 showed activated basal lipolysis and attenuated stimulated lipolysis. Collectively, these findings indicate that PLIN1 regulates lipolysis and adipose homeostasis10,11. In addition, PLIN1 was downregulated in obese WAT, and this resulted in the dysregulation of lipid metabolism including lipolysis12. Several studies have addressed the regulation of PLIN1 expression. For example, the reduced expression of PLIN1 protein in obesity was associated with the promotion of inflammatory responses13. The Plin1 gene is epigenetically regulated, and methylation of its promoter was inversely correlated with basal lipolysis in obese women14. Despite these findings, to date, no consensus has been reached on the mechanisms underlying the obesity-associated downregulation of PLIN1.
Lysosomes are acidic organelles within cells that contain many digestive hydrolases including lipases, phosphatases, and proteases, which degrade specific substrates15,16. Cathepsins are representative lysosomal proteases that play a major role in the degradation of specific proteins17,18. Cathepsins are divided into three groups: aspartyl, cysteine, and serine. The aspartyl cathepsins are represented by cathepsin D (CTSD) and cathepsin E, whereas cysteine cathepsins include cathepsin B (CTSB), and cathepsin L (CTSL), CTSB, CTSL and CTSD are the most abundant lysosomal proteases19. In addition, these cathepsins are involved in the pathogenesis of cancer, neurodegeneration, and metabolic diseases15,18,20. Many studies have reported that the dysfunction of lysosomal cathepsins occur in obese metabolic organs such as WAT and liver, which underlies part of the obesity-related pathology21. Recently, we reported that lysosomal alterations in obese WAT impaired autophagic clearance and were involved in the early pathology of obesity22. In this previous report, we demonstrated that alterations of CTSL maturation, followed by downregulation of CTSL enzymatic activity during the early development of obesity in WAT exacerbated autophagic clearance leading to autophagosome accumulation22. Moreover, complementary activation of CTSB caused by the downregulation of CTSL enhanced inflammasome activation, leading to inflammatory responses in obese WAT22. Therefore, it is important to investigate the influence of cathepsin alterations on obese WAT in detail.
Recently, the degradation of perilipins by lysosomal machinery was reported23,24, and PLIN2 and PLIN3 were reported as targets for chaperone-mediated autophagy (CMA)25. Thus, here we investigated the involvement of lysosomal alterations in the downregulation of PLIN1 in obese WAT.
Results
Downregulation of PLIN1 expression in obese WAT
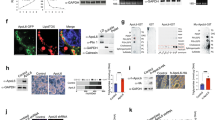
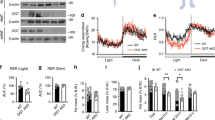
To confirm the dysregulation of PLIN1 in obese WAT, we compared alterations in the expressions of PLIN1 and PLIN2, a perilipin family that is ubiquitously expressed and which participates in LD formation, accompanying high-fat diet (HFD) feeding over a time-course using the normal diet (ND) group as a control. In the 4HFD, 8HFD and 18HFD (HFD intake for 4, 8 and 18 weeks, respectively) groups, a significant decrease in PLIN1 protein expression was observed (Fig. 1A,B). In contrast, a significant increase in PLIN2 (48 kDa) protein expression was found (Fig. 1A,C). Moreover, expression of cell death inducing DFFA like effector c (CIDEC/Fsp27), a marker of LDs and adipocyte differentiation, was unchanged in the 4HFD and 8HFD groups, but decreased in the 18HFD group (Fig. 1A,D). In contrast to protein levels, Plin1 mRNA expression was significantly decreased in the 18HFD group, slightly decreased in the 8HFD group, and unchanged in the 4HFD group (Fig. 1E). In addition, the downregulation of PLIN1 in WAT of the 8HFD group was histologically confirmed (Supplementary Fig. 1A). These results suggest that a decrease in PLIN1 protein preceded that of Plin1 mRNA. Consistent with this finding, immunohistochemical analysis showed increased CTSB protein expression in the WAT of 8HFD mice (Fig. 1F).
Perilipin 1 (PLIN1) protein levels were downregulated in obese WAT. (A) Total protein extracted from WAT of ND, 4HFD, 8HFD and 18HFD mice was analysed by immunoblotting with anti-PLIN1, PLIN2, CIDEC, and GAPDH antibodies (A) and quantified (B–D). Representative images and quantitative data (n = 4) are shown. Intensity of GAPDH was used as a loading control (n = 4). Values indicate mean ± SD. Differences between values were analysed by Student’s t-test with Bonferroni correction. *P < 0.05, **P < 0.01. (E) mRNA expression levels of Plin1 in WAT of ND, 4HFD, 8HFD and 18HFD mice, as analysed by real-time RT-PCR (n = 4). Data were normalized against Tbp (n = 4). Values indicate the mean ± SD. Differences between values were analysed by Student’s t-test with Bonferroni correction. *P < 0.05, **P < 0.01. (F) Immunoenzymatic staining of CTSB in WAT of ND and 8HFD mice. Scale bars represent 200 μm.
Downregulation of PLIN1 is associated with the upregulation of CTSB expression in obese WAT
We recently reported that CTSB protein was significantly increased in 4HFD, 8HFD and 18HFD mice22. These findings suggest that the decrease in PLIN1 protein in early obesity might be associated with an increase in CTSB protein. Generally, it has been reported that macrophage numbers are increased in adipose tissue during obesity and constitute up to 50% of all adipose tissue cells during obesity1,2,3. Therefore, the increased CTSB in obese adipose tissue may be derived from either adipocytes or macrophages. Indeed, we found that CTSB significantly was expressed in macrophages surrounding dead cells, forming so-called “crown-like structures (CLS)” (Fig. 2A). Thus, macrophages that formed CLS showed co-localization with CTSB (Fig. 2A), and CLS was significantly increased in 18HFD mice compared with 8HFD mice (Fig. 2B,C). Taken together, it is likely that the decrease in PLIN1 expression occurs prior to the increase in CLS formation. To clarify whether CTSB localizes in adipocytes or macrophages, we investigated the localization of CTSB protein in ND and 8HFD mice by immunohistochemistry analysis of WAT sections using anti-CTSB and anti-PLIN1 antibodies combined with anti-F4/80 antibody as a marker for macrophages. In agreement with Fig. 1A and previous our work22, a reduction in PLIN1 and an increase in CTSB were observed (Fig. 3A). Simultaneously, part of the CTSB positive area was merged with F4/80, suggesting that CTSB protein was localized to macrophages and adipocytes (Fig. 3A, arrow). In ND mice, the expression of Ctsb mRNA was significantly higher in the adipocyte enrichment fraction (AEF) than the stromal vascular fraction (SVF), and Ctsb expression was increased in both fractions of 8HFD mice (Fig. 3B). Interestingly, we also clearly detected that PLIN1 protein and CTSB protein were alternatively localized in the cytoplasm of adipocytes in the obese WAT of 8HFD mice (Fig. 3A, arrowhead). Therefore, we hypothesized that PLIN1 protein in obese WAT is degraded by increased CTSB in adipocytes at the early stage of obesity, and that increased PLIN1 protein degradation and reduced PLIN1 gene expression contribute to the net protein loss in the late stage of obesity.
Localization of CTSB between adipocytes and macrophages in obese WAT. (A) Representative images of WAT sections from 8HFD mice immunostained for PLIN1, CTSB, and F4/80 (lower panel). Representative images of WAT sections from 8HFD mice immunostained for PLIN1, rabbit-IgG, and rat-IgG (upper panel). Scale bars represent 50 μm. Arrowheads indicate CLS. (B,C) Increased crown-like structures in 18HFD mice. Representative images (B) and quantitative data (CLS per > 300 adipocytes) (C) of WAT sections in ND, 8HFD, and 18HFD mice. Arrowheads indicate CLS. Values indicate the mean ± SD. Differences between values were analysed by Student’s t-test with *P < 0.05, **P < 0.01.
Localization of CTSB in the adipocyte fraction of obese WAT. (A) Representative images of WAT sections from ND and 8HFD mice immunostained for PLIN1, CTSB and F4/80. Scale bars represent 50 μm (upper and middle panels) and 10 μm (lower panels). Arrow indicates the co-localization between CTSB and F4/80, and arrowheads indicate the co-localization between CTSB and adipocytes. (B) qRT-PCR analysis for Ctsb mRNA levels in the adipocyte enriched fraction (AEF) and stromal vascular fraction (SVF) obtained from the WAT of ND and 8HFD groups. Values indicate the mean ± SD. Differences between values were analysed by Tukey-Kramer method with *P < 0.05, **P < 0.01 (n = 4).
CTSB overexpression induces PLIN1 downregulation in 3T3L1 adipocytes
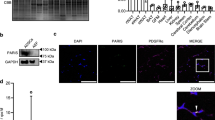
To analyse the relationship between PLIN1 expression and CTSB activity, we generated CTSB-OE 3T3L1 adipocytes. A significant decrease in PLIN1 protein was observed in CTSB-OE cells (Fig. 4A,B). In contrast, PLIN2 and CIDEC were unaltered in CTSB-OE cells (Fig. 4A,C,D). Moreover, treatment with CA074ME, a CTSB inhibitor, whose concentration had no effect on cell viability (Supplementary Fig. 2A)26,27, rescued PLIN1 expression in CTSB-OE cells (Fig. 4E,F). E64d, another cysteine cathepsin inhibitor analogous to CA074ME, also abrogated the CTSB OE-induced reduction in PLIN1 expression, while pepstatin A, an aspartyl cathepsin inhibitor that suppresses the activity of CTSD and CTSE, failed to exert a similar effect (Supplementary Fig. 2B). However, the mRNA expression of Plin1 and Plin2 did not change in CTSB-OE cells with or without CA074ME (Supplementary Fig. 2C,D). These results indicate that CTSB may be associated with reduced PLIN1 protein expression. Immunofluorescent analysis showed that CTSB overexpression further reduced the PLIN1 signal around LDs (Fig. 4G). Similar to the results shown in Fig. 4E,F, this reduction was reversed by treatment with CA074ME (Fig. 4G). These data indicate that CTSB overexpression induced the elimination of PLIN1 protein.
Degradation of PLIN1 by CTSB. (A–D) Total cell lysates extracted from Mock or CTSB-OE 3T3L1 adipocytes were analysed by immunoblotting using anti-PLIN1, PLIN2, CIDEC, FLAG, and LMNB1 antibodies (A) and quantified (B–D). Representative images and quantitative data (n = 4) are shown. Intensity of LMNB1 was used as a loading control. Values indicate the mean ± SD. Differences between values were analysed by Student’s t-test with *P < 0.05, **P < 0.01. (E,F) Mock or CTSB-OE 3T3L1 adipocytes were treated with 10 μM CA074ME for 24 h. Total cell lysates were analysed by immunoblotting using anti-PLIN1, CIDEC, and LMNB1 antibodies (E) and quantified (F). Representative images and quantitative data (n = 4) are shown. Intensity of LMNB1 was used as a loading control. Values indicate the mean ± SD. Differences between values were analysed by Tukey-Kramer method with *P < 0.05, **P < 0.01. (G) Mock or CTSB-OE 3T3L1 adipocytes were treated with 10 μM CA074ME for 24 h. Immunofluorescence analysis was performed with an anti-PLIN1 antibody. PLIN1 (green), lipid droplets (red) and nucleus (blue) are shown. Data shown are representative of individual experiments. Scale bars represent 20 μm.
Because PLIN1 protein expression is regulated by peroxisome proliferator activated receptor γ (PPARG), a master regulator of adipogenesis28, we considered that the reduced PLIN1 observed in CTSB-OE cells might be attributed to the dysregulation of adipogenesis. To rule out this possibility, we investigated the degree of adipogenesis in CTSB-OE cells and found that triacylglycerol accumulation was significantly increased in CTSB-OE cells (Supplementary Fig. 3A,B). In addition, the expressions of PPARG1 and PPARG2 proteins were more robust over days 2 to 4 during adipocyte differentiation in CTSB-OE cells than in control cells (Supplementary Fig. 3C–E). Moreover, although the expression of CIDEC was unchanged, PLIN1 protein was significantly decreased during adipocyte differentiation in CTSB-OE cells (Supplementary Fig. 3C,F,G). These results indicate accelerated adipogenesis in CTSB-OE cells, in agreement with a previous report29. Thus, we concluded that reduced PLIN1 protein in CTSB-OE cells occurs independent of changes in adipogenesis.
Localization of CTSB and PLIN1 in 3T3L1 adipocytes
To determine the localization of CTSB protein in adipocytes, we generated 3T3L1 cells expressing CTSB-mCherry fusion protein, because commercially available CTSB antibodies failed to work for cellular immunostaining in our experiments. In the differentiated CTSB-mCherry-OE adipocytes, most exogenous mCherry was colocalized with lysosomal membrane protein LAMP2, indicating the lysosomal localization of CTSB protein in adipocytes (Fig. 5A,B). Next, to investigate the association between lysosomes and PLIN1, we analysed the localization of LAMP2 and PLIN1 in differentiated 3T3-L1 cells treated with a CTSB inhibitor to facilitate the detection of PLIN1. LAMP2 was in contact with the PLIN1 protein, suggesting that the lysosome interacts with PLIN1 on LDs (Fig. 5C). Furthermore, we identified that CTSB-mCherry was also in contact with PLIN1 (Fig. 5D). These results suggest that PLIN1 protein was associated with CTSB on LDs.
Localization of PLIN1 in CTSB-OE 3T3L1 adipocytes. (A and B) Immunofluorescence analysis was performed with anti-LAMP2 antibodies in CTSB-mCherry-OE 3T3L1 adipocytes. Images of CTSB-mCherry (red) and LAMP2 (green) (A), and histogram of fluorescence intensity (B) are shown. Representative images of individual experiments are shown. Scale bar represents 10 μm. Asterisk indicates LD. (C) CTSB-OE 3T3L1 adipocytes were treated with 10 μM CA074ME for 24 h. Immunofluorescence analysis was performed with anti-PLIN1 and LAMP2 antibodies. PLIN1 (green) and LAMP2 (red) are shown. Representative images of individual experiments are shown. Scale bar represents 5 μm. Arrows indicate the contact site between PLIN1 and LAMP2. (D) CTSB-mCherry-OE 3T3L1 adipocytes were treated with 10 μM CA074ME for 24 h. Immunofluorescence analysis was performed with an anti-PLIN1 antibody. PLIN1 (green) and CTSB-mCherry (red) are shown. Representative images of individual experiments are shown. Scale bar represents 5 μm. Arrows indicate the contact site between PLIN1 and CTSB-mCherry.
CTSB overexpression disturbs lipid metabolism
To evaluate the implications of CTSB overexpression-induced PLIN1 downregulation on basal lipolysis, we examined lipolysis, which was evaluated by the release of glycerol into the medium, and intracellular triacylglycerol (TG) content in CTSB-OE cells with or without CA074ME treatment. Initially, we investigated the size of LD in Mock cells and CTEB-OE cells, but failed to detect any differences (Fig. 6A,B). However, CTSB overexpression enhanced the release of glycerol, which was rescued by CA074ME treatment (Fig. 6C). These results indicated that CTSB overexpression induced futile basal lipolysis because of the PLIN1 degradation. Interestingly, CTSB-OE cells exhibited increased intracellular TG content compared with Mock cells, which was inhibited by CA074ME treatment (Fig. 6D). Such phenotypic changes in TG content were associated with increased lipid metabolism30,31. Therefore, CTSB overexpression is associated with the alteration of lipid metabolism in adipocytes.
Dysregulation of lipid metabolism by CTSB overexpression in 3T3L1 adipocytes. (A and B) Lipid droplets (LDs) were stained by Nile red in Mock or CTSB-OE 3T3L1 adipocytes. Representative images (A) and quantitative data of the mean LD diameter (B) are shown. Scale bar represents 50 μm. Values indicate the mean ± SD. Differences between values were analysed by Student’s t-test with *P < 0.05, **P < 0.01. (C and D) Release assays for glycerol (C) and measurement of intracellular TG content (D) were performed in Mock or CTSB-OE 3T3L1 adipocytes. Values indicate the mean ± SD. Differences between values were analysed by Tukey-Kramer method with *P < 0.05, **P < 0.01.
Discussion
Several studies have demonstrated that obesity reduces PLIN1 expression. For example, Gaidhu et al. found that downregulated PLIN1 protein expression in obese WAT caused lipolytic dysfunction in obese WAT12. However, the underlying mechanism remains incompletely understood. It was reported that PLIN1 mRNA expression was lower in WAT from obese individuals compared with that from non-obese subjects32. Conversely, recent studies have demonstrated that PLIN1 protein is degraded by lysosomes23,24,33. In addition, Kovsan et al. showed significantly increased PLIN1 protein expression in adipocytes treated with the lysosomal protease inhibitor leupeptin, but not the lysosomal acidity inhibitor NH4Cl, suggesting that PLIN1 protein may be degraded specifically by cathepsin23. Leupeptin inhibits sulfhydryl proteases such as plasmin, papain, trypsin, and CTSB34,35. Therefore, we hypothesized that CTSB contributes to the degradation of PLIN1 protein in obese WAT. In the present study, we demonstrated that CTSB overexpression led to reduced PLIN1 protein and altered lipid metabolism in 3T3L1 adipocytes. Furthermore, a decrease in PLIN1 protein levels occurred in the early stage of HFD (4HFD), but Plin1 mRNA levels failed to significantly decrease until the late stage of HFD (18HFD). Therefore, we speculated the decrease of PLIN1 protein in 4HFD and 8HFD may be mainly related to the upregulation of CTSB activity, whereas in 18HFD, the downregulation of Plin1 mRNA levels might synergistically contribute to decreased PLIN1 protein in addition to protein degradation by CTSB. Considering a previous study reporting that the Plin1 gene is epigenetically regulated14, we speculated that Plin1 mRNA changes might be related to epigenetic regulation in the late stage of obesity. Furthermore, we observed that PLIN2 protein abundance was increased in obese WAT unlike PLIN1. It was reported that PLIN2 proteins accumulated in CLS macrophages in obese WAT36. Therefore, given our result that PLIN2 protein is unchanged in CTSB-OE cells, it is likely that the obesity-induced upregulation of PLIN2 in WAT may be attributed to the increased infiltration of macrophages, rather than CTSB overexpression in obese adipocytes.
In addition to CTSB, we also identified that CTSD, a lysosomal serine protease ubiquitously expressed in various cells/tissues, was upregulated in obese WAT (Supplementary Fig. 4A). Therefore, to clarify the relationship between CTSD and PLIN1, we used CTSD-OE 3T3L1 adipocytes and examined the effects on PLIN1 expression (Supplementary Fig. 4B). CTSD overexpression, however, did not affect PLIN1 protein levels (Supplementary Fig. 4C–E). These results suggest that CTSD, a major lysosomal hydrolase, does not contribute to the degradation of PLIN1. It was reported that CTSB directly degraded tenascin-C, an extracellular matrix glycoprotein37,38. We showed here that CTSB was involved in the reduction of PLIN1 in obese WAT. Therefore, it is likely that PLIN1 is a novel-selective substrate of CTSB.
Regarding how CTSB and PLIN interact with each other, we revealed that exogenous CTSB protein was mainly localized in lysosomes in CTSB-OE cells and in contact with PLIN1 protein on LDs (Fig. 5). We propose two mechanisms for this phenomenon as follows. One possibility involves chaperone-mediated autophagy (CMA), which selectively degrades a subset of cytosolic proteins by direct sequestering in lysosomes. As stated above, both PLIN2 and PLIN3 proteins are targets of CMA25. Because the perilipin family shares 43% amino acid sequence identity4, PLIN1 protein might also be a target of CMA. A second possibility is that CTSB released by lysosomal membrane permeabilization (LMP) interacts with PLIN1 in cytoplasm39,40. Several factors, such as oxidative stress, fatty acids, and lysosomotropic compounds, induce LMP and cause the release of cathepsins and other hydrolases from the lysosomal lumen into the cytosol39. This release occurs under conditions of cell death including apoptosis or necrosis39. In obese WAT, PLIN1 protein is not detected by immunohistochemistry during adipocyte cell death, which is generally characterized by increased CLS36. These reports support the notion that CTSB released by LMP causes the degradation of the PLIN1 protein in obese WAT. However, our results also indicate that decreased PLIN1 expression occurs prior to the increase in CLS (Figs. 1–3). Thus, we suggest that CMA and the release of CTSB accompanying LMP might play a role in PLIN1 degradation at the early and advanced stages of obesity, respectively. In the future, it will be important to examine further the involvement of CMA and LMP in the degradation of PLIN1.
PLIN1 has multiple important phosphorylation sites (Ser-434, Ser-492, and Ser-517) in its C-terminal region that regulate its function4. It is generally accepted that lipolysis is dysregulated in the WAT of obese mice and obese humans, attributable to decreased PLIN1 and HSL protein expressions12,32,41,42. Moreover, in PLIN1-knockout mice, basal lipolysis was induced, but stimulated lipolytic activity was attenuated11. PLIN1 knockdown in 3T3-L1 adipocytes dramatically increased basal lipolysis43. Our results also showed that the CTSB-induced degradation of PLIN1 promoted basal lipolysis. These findings imply that CTSB overexpression causes dysfunctional basal lipolysis through PLIN1 downregulation in obese WAT. However, CTSB-OE cells displayed accelerated adipocyte differentiation and increased TG content, the latter of which was suppressed by CA074ME, implying that CTSB may directly regulate lipogenesis separate from adipocyte differentiation. A previous study showed that lipolysis and lipogenesis were regulated in parallel in adipocytes during the remodelling of LDs30,31, which supports the idea that increased TG levels in CTSB-OE cells might occur independently of alterations in lipolysis. Therefore, although hypertrophic adipocytes with increased TG content were suspected to release more glycerol, we assumed that CTSB overexpression contributed to increased basal lipolysis via reduced PLIN1 and lipogenesis via unknown mechanisms. It is widely accepted that FFAs released from hypertrophic adipocytes activate macrophages primarily via the Toll-like receptor 4-JNK pathway and induce proinflammatory signals13,44,45. Therefore, the upregulation of basal FFA released via CTSB-induced PLIN1 degradation in adipocytes might contribute to a chronic state of low-grade inflammation, which is usually observed in obese WAT, leading to a pathological state of obesity.
In conclusion, our data provide the first evidence that CTSB overexpression in obese WAT is associated with reduced PLIN1 protein expression, resulting in abnormal futile basal lipolysis and a possible subsequent inflammatory response. This study proposes an important mechanism of PLIN1 regulation and the possibility that CTSB might be a novel therapeutic target for obesity.
Materials and Methods
Animals and diets
All animal experiments and protocols were conducted in accordance with the Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology of Japan, and were approved by the Ethics Review Committee for Animal Experimentation at the Tokyo University of Science (approval number Y14028). Male C57BL/6 mice (3 weeks of age) were purchased from CREA Japan (Tokyo, Japan) and maintained in temperature-controlled, specific pathogen-free conditions with 12-h light/dark cycles within the animal facility at Tokyo University of Science. For experiments, male C57BL/6 mice (4 weeks of age) were randomly divided into four groups: normal diet (ND; Nosan, Yokohama, Japan), 4HFD, 8HFD and 18HFD. The latter three groups were fed a high-fat diet (HFD32, CREA) for 4, 8 or 18 weeks, respectively, before euthanasia, whereas ND mice were fed a ND throughout their lives. After 22 weeks, all four groups of mice were euthanized and epididymal WAT was collected.
Cell culture and treatment
3T3-L1 pre-adipocytes were purchased from RIKEN Bioresource Center (Ibaraki, Japan) and maintained in Dulbecco’s Modified Eagle’s Medium (low glucose; 041–29775, Wako, Osaka, Japan) with 10% foetal bovine serum and 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA). Differentiation of 3T3-L1 pre-adipocytes to adipocytes (Day 8) was performed as previously reported by our laboratory46,47. Pepstatin A1 (4397-v) and CA074ME (4323-v) were purchased from Peptide Institute (Osaka, Japan).
Retrovirus vector construction and infection of 3T3L1 pre-adipocytes
Retrovirus-CTSB overexpression vector and retrovirus-CTSB-mCherry were constructed according to a previous report22. Briefly, CTSB cDNA was amplified from a CTSB vector (11249, Addgene, Cambridge, MA, USA) by PCR using PrimeSTAR HS DNA polymerase (R010A, Takara, Kyoto, Japan), and then subcloned into a Xho1- and Not1-digested pMXs-AMNN-Puro vector46,47. Oligonucleotide primers for CTSB were chemically synthesized. CTSD cDNA was amplified from mouse cDNA libraries using the chemically synthesized primers. For the CTSB-mCherry vector, CTSB cDNA was subcloned into a Xho1- and HindIII-digested pmCherry-N1 vector. Subcloned vectors were then digested with Xho1 and Not1 and cloned into a Xho1- and Not1-digested pMXs-AMNN-Puro vector46,47. The sequences of used primers are shown in the primer list (Supplementary Fig. 5A). Retroviral vectors were produced by transfecting Plat-E cells with the resulting vector plasmids, as described previously48. The retrovirus particles were concentrated with a 4 × PEG-it solution (32% (w/v) PEG-6000, 400 mM NaCl, and 40 mM HEPES, pH 7.4). To obtain 3T3L1 cell lines overexpressing CTSB, CTSD, or CTSB-mCherry, 3T3L1 cells were incubated with virus-containing medium for 2 days and then selected with 2 μg/mL puromycin for 5 days.
Immunoblotting analysis
Immunoblotting with Immunostar LD chemiluminescent substrates (290–69904, Wako) was performed. Signals were detected with an LAS-3000 image analyser (Fujifilm, Tokyo, Japan) as previously described49,50. Lamin β1 (LMNB1; PM064, 1:10000) was purchased from MBL (Nagoya, Japan). CTSB (ab58802, 1:400), CIDEC/FSP27 (ab16760, 1:2000), PLIN1 (ab61682, 1:400) and PLIN2 (ab108323, 1:1000) were purchased from Abcam (Cambridge, UK). FLAG M2 (F1804, 1:5000) was purchased from Sigma-Aldrich. PPARG (sc-7273, 1:2000) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (010-25521, 1:5000) was purchased from Wako.
Cathepsin activity assay
Cathepsin activity was assayed as previously reported22,51. Briefly, fluorometric analysis with Z-Arg-Arg-MCA (3123-v, Peptide Institute) for CTSB and MOCAc-Gly-Lys-Pro-Ile-Leu-Phe-Phe-Arg-Leu-Lys(Dnp)-D-Arg-NH2 for CTSD (3200-v, Peptide Institute) was performed as previously reported22 using WAT or cell pellets resuspended in lysis buffer (352 mM KH2PO4, 48 mM Na2HPO4, 4 mM EDTA, pH 6.0) and incubated on ice for 60 min before centrifugation for 10 min at 2100 × g. Supernatant was collected and protein concentrations were determined with a BCA kit (23225, Pierce, Waltham, MA, USA). Supernatant was then added to the reaction buffer (4 mM DTT in lysis buffer) as an assay buffer.
For the measurement of CTSB activity, 100 μL assay buffer (containing 1 μg of protein) was mixed with 100 μL of substrate buffer (10 μM Z-Arg-Arg-AMC diluted in 0.1% Brij 35; B4184, Sigma-Aldrich) and incubated at 37 °C for 30 min. Fluorescence was measured using a Wallac ARVO MX/Light 1420 Multilabel/Luminescence Counter (PerkinElmer, Waltham, MA, USA) with an excitation/emission of 360/460 nm.
For the measurement of CTSD activity, 100 μL of assay buffer (containing 1 μg of protein) was mixed with 100 μL of substrate buffer (10 μM CTSD substrate diluted in 0.1% Brij 35) and incubated at 37 °C for 30 min. Fluorescence was measured using a Wallac ARVO MX/Light 1420 Multilabel/Luminescence Counter with an excitation/emission of 360/460 nm.
Quantitative real-time RT-PCR
Total RNA was extracted from frozen WAT using the ReliaPrep RNA Tissue Miniprep System (Promega, Tokyo, Japan), in accordance with the manufacturer’s protocol. To obtain cDNA, 1 μg of RNA was subjected to reverse transcription using PrimeScript Reverse Transcriptase (Takara, Shiga, Japan) and random hexamer primers (Takara). Quantitative real-time RT-PCR was performed using a CFX Connect Real-Time PCR System (Bio-Rad, Hercules, CA, USA) with Thunder Bird SYBR qPCR Mix (QPS-201, Osaka, Japan), as previously described52. The sequences of used primers are shown in the primer list (Supplementary Fig. 5B).
Immunofluorescence and confocal microscopy in 3T3L1 adipocytes
3T3L1/Mock, 3T3L1/CTSB-overexpressing (OE), and 3T3L1/CTSB-mCherry-OE pre-adipocytes grown on cover slips were differentiated into adipocytes and treated with or without 10 μM of the cathepsin B inhibitor CA074ME (4323-v, Peptide Institute) for 24 h. After fixing with 4% paraformaldehyde for 15 min, cells were permeabilized and incubated with primary antibodies against PLIN1 (ab61682, 1:400, Abcam) or LAMP2 (ab13524, 1:1000, Abcam) at 4 °C overnight, and then with Alexa Fluor-conjugated secondary antibodies (Alexa Fluor-488 goat anti-rabbit IgG (H + L), A11070, 1:1000, Invitrogen, Carlsbad, CA, USA; Alexa Fluor-594 goat anti-rabbit IgG (H + L), A110121, Invitrogen; Alexa Fluor-488 goat anti-rat IgG (H + L), ab150157, Abcam) for 1 h at room temperature. Hoechst 33342 (H3570, 1:10000, Invitrogen) and Nile red were used to stain nuclei and LDs, respectively. Images were acquired on an SP8 confocal microscope (Leica, Wetzlar, Germany) at 63 × magnification.
Immunohistochemistry of WAT
WAT samples from ND and 8HFD mice were fixed with 10% formalin for 24 h and embedded in paraffin. Sections (5 μm thick) were cut from each tissue block, deparaffinized, and dehydrated. Antigen retrieval was performed on samples by heating in 10 mM citrate buffer solution (pH 6.0) supplemented with 0.1% NP40 in an autoclave for 10 min at 121 °C. After blocking of nonspecific reactivity with bovine serum albumin and goat serum for 60 min at room temperature, sections were incubated overnight at 4 °C with anti-PLIN1 (GP29, 1:200, Progen Biotechnik, Heidelberg, Germany), anti-F4/80 (ab6640, 1:200, Abcam) and anti-CTSB (#31718, 1:200, Cell Signaling Technology, MA, USA) antibodies. As a negative control, sections were incubated with normal rabbit IgG (sc-2027, Santa Cruz Biotechnology) or normal rat IgG (sc-2026, Santa Cruz Biotechnology). Distribution of the primary antibody was achieved by subsequent application of Alexa Fluor-conjugated secondary antibodies (Alexa Fluor-488 goat anti-rat IgG (H + L), A-11006, 1:1000, Invitrogen; Alexa Fluor-555 goat anti-rabbit IgG (H + L), A27039, Invitrogen; Alexa Fluor-405 goat anti-guinea pig IgG (H + L), ab175678, Abcam) for 1 h at room temperature. Representative images were acquired with a confocal microscope (SP8, Leica) at 63 × oil magnification.
Immunohistochemical analysis of CTSB protein expression was performed following the manufacturer’s protocol for the Histofine SAB-PO kit (424032, Nichirei Bioscience, Tokyo, Japan). Briefly, antigen retrieval was performed on samples as described above. Next, endogenous peroxidase was blocked by incubating sections in 0.3% H2O2 in dH2O for 30 min. After blocking of nonspecific reactivity with goat serum for 60 min at room temperature, sections were incubated overnight at 4 °C with an anti-CTSB (sc-13985, 1:5000, Santa Cruz Biotechnology) antibody. As a negative control, sections were incubated with rabbit IgG (2729, Cell Signaling Technology). Distribution of the primary antibody was achieved by subsequent application of a biotinylated anti-rabbit IgG antibody for 1 h at room temperature, followed by incubation for 5 min with peroxidase-labelled streptavidin. Immunostaining was developed using DAB solution (425011, Histofine DAB Substrate Kit, Nichirei Bioscience). Representative images were acquired with a fluorescence microscope (DMi8, Leica) at 20 × magnification.
Light microscopy and morphometry
WAT sections (5 μm thick) were stained with haematoxylin and eosin to assess morphology. Images were acquired with a light microscope (Eclipse 80i; Nikon, Tokyo, Japan) using a 10 × objective lens. CLS density (CLS per more than 300 adipocytes) was quantified by ImageJ software (https://imagej.nih.gov/ij/).
Glycerol release assay and intracellular TG content
A glycerol release assay (Adipolysis assay kit; Cayman Chemical, Ann Arbor, MI, USA) with medium from differentiated 3T3-L1/Mock and 3T3L1/CTSB-OE adipocytes and intracellular TG content (LabAssay™ Triglyceride, Wako) was performed according to the manufacturer’s protocols. Glycerol content in the media was measured calorimetrically using a set of standards. Cells were then washed with cold PBS, lysed in 2% SDS buffer, and the protein concentration was determined and used to normalize glycerol release. All experiments were carried out in triplicate.
Statistical analyses
Statistical analyses were performed using the Tukey–Kramer test (with R software version 3.1.0) or Student’s t-test (with or without Bonferroni correction). Data are presented as the mean ± standard deviation. P-values < 0.05 were considered statistically significant.
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
References
Furukawa, S. et al. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Invest. 114, 1752–1761 (2004).
Cinti, S. The adipose organ. Prostaglandins. Leukot. Essent. Fatty. Acids. 73, 9–15 (2005).
Ouchi, N., Parker, J. L., Lugus, J. J. & Walsh, K. Adipokines in inflammation and metabolic disease. Nat. Rev. Immunol. 11, 85–97 (2011).
Brasaemle, D. L. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid. Res. 48, 2547–2559 (2007).
Wang, H. & Sztalryd, C. Oxidative tissue: perilipin 5 links storage with the furnace. Trends. Endocrinol. Metab. 22, 197–203 (2011).
Konige, M., Wang, H. & Sztalryd, C. Role of adipose specific lipid droplet proteins in maintaining whole body energy homeostasis. Biochim. Biophys. Acta. 1842, 393–401 (2014).
Miyoshi, H. et al. Perilipin promotes hormone-sensitive lipase-mediated adipocyte lipolysis via phosphorylation-dependent and -independent mechanisms. J. Biol. Chem. 281, 15837–15844 (2006).
Marcinkiewicz, A., Gauthier, D., Garcia, A. & Brasaemle, D. L. The phosphorylation of serine 492 of perilipin a directs lipid droplet fragmentation and dispersion. J. Biol. Chem. 281, 11901–11909 (2006).
Miyoshi, H. et al. Control of adipose triglyceride lipase action by serine 517 of perilipin A globally regulates protein kinase A-stimulated lipolysis in adipocytes. J. Biol. Chem. 282, 996–1002 (2007).
Martinez-Botas, J. et al. Absence of perilipin results in leanness and reverses obesity in Lepr(db/db) mice. Nat. Genet. 26, 474–479 (2000).
Tansey, J. T. et al. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc. Natl. Acad. Sci. USA 98, 6494–6499 (2001).
Gaidhu, M. P. et al. Dysregulation of lipolysis and lipid metabolism in visceral and subcutaneous adipocytes by high-fat diet: role of ATGL, HSL, and AMPK. Am. J. Physiol. Cell. Physiol. 298, C961–971 (2010).
Sohn, J. H. et al. Perilipin 1 (Plin1) deficiency promotes inflammatory responses in lean adipose tissue through lipid dysregulation. J. Biol. Chem. 293, 13974–13988 (2018).
Bialesova, L. et al. Epigenetic Regulation of PLIN 1 in Obese Women and its Relation to Lipolysis. Sci. Rep. 7, 10152 (2017).
Appelqvist, H., Wäster, P., Kågedal, K. & Öllinger, K. The lysosome: from waste bag to potential therapeutic target. J. Mol. Cell. Biol. 5, 214–226 (2013).
Settembre, C., Fraldi, A., Medina, D. L. & Ballabio, A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell. Biol. 14, 283–296 (2013).
Turk, B., Turk, D. & Turk, V. Lysosomal cysteine proteases: more than scavengers. Biochim. Biophys. Acta. 1477, 98–111 (2000).
Olson, O. C. & Joyce, J. A. Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat. Rev. Cancer. 15, 712–729 (2015).
Kaminskyy, V. & Zhivotovsky, B. Proteases in autophagy. Biochim. Biophys. Acta. 1824, 44–50 (2012).
Stoka, V., Turk, V. & Turk, B. Lysosomal cathepsins and their regulation in aging and neurodegeneration. Ageing. Res. Rev. 32, 22–37 (2016).
Mizunoe, Y. et al. Association between Lysosomal Dysfunction and Obesity-Related Pathology: A Key Knowledge to Prevent Metabolic Syndrome. Int. J. Mol. Sci. 20, pii: E3688 (2019).
Mizunoe, Y. et al. Involvement of lysosomal dysfunction in autophagosome accumulation and early pathologies in adipose tissue of obese mice. Autophagy. 13, 642–653 (2017).
Kovsan, J., Ben-Romano, R., Souza, S. C., Greenberg, A. S. & Rudich, A. Regulation of adipocyte lipolysis by degradation of the perilipin protein: nelfinavir enhances lysosome-mediated perilipin proteolysis. J. Biol. Chem. 282, 21704–21711 (2007).
Ogasawara, J. et al. Oligonol-induced degradation of perilipin 1 is regulated through lysosomal degradation machinery. Nat. Prod. Commun. 7, 1193–1196 (2012).
Kaushik, S. & Cuervo, A. M. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell. Biol. 17, 759–770 (2015).
ISO 10993-5:2009 Biological Evaluation of Medical Devices. Part 5: Tests for In Vitro Cytotoxicity. International Organization for Standardization; Geneva, Switzerland (2009).
López-García, J., Lehocký, M., Humpolíček, P. & Sáha, P. HaCaT Keratinocytes Response on Antimicrobial Atelocollagen Substrates: Extent of Cytotoxicity, Cell Viability and Proliferation. J Funct Biomater. 5, 43–57 (2014).
Arimura, N., Horiba, T., Imagawa, M., Shimizu, M. & Sato, R. The peroxisome proliferator-activated receptor gamma regulates expression of the perilipin gene in adipocytes. J. Biol. Chem. 279, 10070–10076 (2004).
Zhang, Z. Y. et al. CTSB promotes porcine preadipocytes differentiation by degrading fibronectin and attenuating the Wnt/β-catenin signaling pathway. Mol. Cell. Biochem. 395, 53–64 (2014).
Paar, M. et al. Remodeling of lipid droplets during lipolysis and growth in adipocytes. J Biol Chem. 287, 11164–11173 (2012).
Ali, S. B., Turner, J. J. O. & Fountain, S. J. Constitutive P2Y2 receptor activity regulates basal lipolysis in human adipocytes. J Cell Sci. 131, pii: jcs221994 (2018).
Wang, Y. et al. Perilipin expression in human adipose tissues: effects of severe obesity, gender, and depot. Obes. Res. 11, 930–936 (2003).
Xu, G., Sztalryd, C. & Londos, C. Degradation of perilipin is mediated through ubiquitination-proteasome pathway. Biochim Biophys Acta. 1761, 83–90 (2006).
Furuno, K., Ishikawa, T. & Kato, K. Appearance of autolysosomes in rat liver after leupeptin treatment. J. Biochem. 91, 1485–1494 (1982).
Glaumann, H. & Ahlberg, J. Comparison of different autophagic vacuoles with regard to ultrastructure, enzymatic composition, and degradation capacity–formation of crinosomes. Exp Mol. Pathol. 47, 346–362 (1987).
Cinti, S. et al. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J. Lipid. Res. 46, 2347–2355 (2005).
Fonović, M. & Turk, B. Cysteine cathepsins and extracellular matrix degradation. Biochim. Biophys. Acta. 1840, 2560–2570 (2014).
Mai, J., Sameni, M., Mikkelsen, T. & Sloane, B. F. Degradation of extracellular matrix protein tenascin-C by cathepsin B: an interaction involved in the progression of gliomas. Biol. Chem. 383, 1407–1413 (2002).
Boya, P. & Kroemer, G. Lysosomal membrane permeabilization in cell death. Oncogene. 27, 6434–6451 (2008).
Serrano-Puebla, A. & Boya, P. Lysosomal membrane permeabilization in cell death: new evidence and implications for health and disease. Ann. N Y. Acad. Sci. 1371, 30–44 (2016).
Langin, D. et al. Adipocyte lipases and defect of lipolysis in human obesity. Diabetes. 54, 3190–3197 (2005).
Díaz-Ruiz, A. et al. Proteasome Dysfunction Associated to Oxidative Stress and Proteotoxicity in Adipocytes Compromises Insulin Sensitivity in Human Obesity. Antioxid. Redox. Signal. 23, 597–612 (2015).
Sun, Z. et al. Perilipin1 promotes unilocular lipid droplet formation through the activation of Fsp27 in adipocytes. Nat. Commun. 4, 1594, https://doi.org/10.1038/ncomms2581 (2003).
Nguyen, M. T. et al. A subpopulation of macrophages infiltrates hypertrophic adipose tissue and is activated by free fatty acids via Toll-like receptors 2 and 4 and JNK-dependent pathways. J. Biol. Chem. 282, 35279–35292 (2007).
Glass, C. K. & Olefsky, J. M. Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab. 15, 635–645 (2012).
Kaneko, H. et al. Taurine is an amino acid with the ability to activate autophagy in adipocytes. Amino Acids. 50, 527–535 (2018).
Okita, N. et al. Inhibitory effect of p53 on mitochondrial content and function during adipogenesis. Biochem. Biophys. Res. Commun. 446, 91–97 (2014).
Kobayashi, M. et al. Functional Analysis of Novel Candidate Regulators of Insulin Secretion in the MIN6 Mouse Pancreatic β Cell Line. PLoS. One. 11, e0151927, https://doi.org/10.1371/journal.pone.0151927 (2016).
Kobayashi, M. et al. Mitochondrial intermediate peptidase is a novel regulator of sirtuin-3 activation by caloric restriction. FEBS Lett. 591, 4067–4073 (2017).
Mizunoe, Y. et al. Trehalose protects against oxidative stress by regulating the Keap1-Nrf2 and autophagy pathways. Redox Biol. 15, 115–124 (2017).
Barrett, A. J. & Kirschke, H. CTSB, cathepsin H, and CTSL. Methods Enzymol. 80, Pt C:535–561 (1981).
Fujii, N. et al. Sterol regulatory element-binding protein-1c orchestrates metabolic remodeling of white adipose tissue by caloric restriction. Aging. Cell. 16, 508–517 (2017).
Acknowledgements
We thank all members of the Laboratory of Molecular Pathology and Metabolic Disease of Faculty of Pharmaceutical Sciences, Tokyo University of Science (TUS) for their cooperation. The authors also thank Drs. Hitoshi Shimano, Yoshimi Nakagawa, University of Tsukuba, for their help. We also thank Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript. Y.M. was supported by a Grant-in-Aid for Early-Career Scientists (No. 18K17953) from the Japan Society for the Promotion of Science. M.K. was supported by a Grant-in-Aid for Young Scientists (B) (No. 17K13231) from the Japan Society for the Promotion of Science. Y.H. was supported by a Grant-in-Aid for Scientific Research (B) (No. 17H02179) from the Japan Society for the Promotion of Science, and by the MEXT Supported Program for the Strategic Research Foundation at Private Universities, 2014–2018.
Author information
Authors and Affiliations
Contributions
Y.H. conceived the idea, designed experiments, and wrote the manuscript for this study. Y.M., M.K. and S.H. designed and performed experiments, and Y.M. and M.K. also wrote the manuscript for this study. Y.S., N.O., A.H., R.I., R.T., Y.N., R.T. and H.S. supported the experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mizunoe, Y., Kobayashi, M., Hoshino, S. et al. Cathepsin B overexpression induces degradation of perilipin 1 to cause lipid metabolism dysfunction in adipocytes. Sci Rep 10, 634 (2020). https://doi.org/10.1038/s41598-020-57428-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-57428-6
This article is cited by
-
Lipid droplets: a cellular organelle vital in cancer cells
Cell Death Discovery (2023)
-
A Brain-Penetrant Stearoyl-CoA Desaturase Inhibitor Reverses α-Synuclein Toxicity
Neurotherapeutics (2022)
-
Novel Targets in Glucose Homeostasis and Obesity—Lesson from Rare Mutations
Current Diabetes Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.