Abstract
Glioblastoma multiforme (GBM) is characterized by highly invasive growth, which leads to extensive infiltration and makes complete tumor excision difficult. Since cytoskeleton proteins are related to leading processes and cell motility, and through analysis of public GBM databases, we determined that an actin-interacting protein, zyxin (ZYX), may involved in GBM invasion. Our own glioma cohort as well as the cancer genome atlas (TCGA), Rembrandt, and Gravendeel databases consistently showed that increased ZYX expression was related to tumor progression and poor prognosis of glioma patients. In vitro and in vivo experiments further confirmed the oncogenic roles of ZYX and demonstrated the role of ZYX in GBM invasive growth. Moreover, RNA-seq and mass-spectrum data from GBM cells with or without ZYX revealed that stathmin 1 (STMN1) was a potential target of ZYX. Subsequently, we found that both mRNA and protein levels of STMN1 were positively regulated by ZYX. Functionally, STMN1 not only promoted invasion of GBM cells but also rescued the invasion repression caused by ZYX loss. Taken together, our results indicate that high ZYX expression was associated with worse prognosis and highlighted that the ZYX-STMN1 axis might be a potential therapeutic target for GBM.
Similar content being viewed by others
Introduction
Gliomas are the most common primary tumors of the central nervous system in children and adults, of which GBM multiforme (GBM) is the most malignant form and classified as a grade IV glioma according to the World Health Organization (WHO) system [1]. GBM is featured by highly invasive growth and poor survival, with a median survival time about 1 year from initial diagnosis [2]. GBM cells are notorious for their invasiveness, which leads to its high recurrence rate and reduced life expectancy [3, 4]. Due to the highly invasive growth, it is almost impossible to completely remove the tumor mass through regular surgical procedures. Therefore, identifying reliable biomarkers for invasiveness of GBM is crucial for prediction of patient prognosis as well as for identification of novel therapeutic drug targets.
Migration and invasion are critically dependent on dynamic organization of the cytoskeleton. Generally, the cytoskeleton governs the cell size, shape, migration, proliferation, through which the cells respond to their environment. Mutations and abnormal expression of cytoskeletal and cytoskeletal-associated proteins play important roles in tumorigenesis and tumor progression, including tumor cell migration, invasion, and metastasis. It is well known that migration and invasion mediated by the cytoskeleton require functional focal adhesion and the interaction between the cell and extracellular substrates [5,6,7]. By combing datasets related to cell–substrate interaction from Gene Ontology and the dataset on focal adhesion from KEGG, followed by analysis of public GBM databases, we identified ZYX as a potential critical candidate involved in GBM invasive growth.
In previous work, the effect of ZYX on the invasiveness of tumor cells has been reported in other types of cancers. For example, Yamamura et al. determined that ZYX promotes invasiveness of oral squamous cell carcinoma cells by upregulating Rac1 and Cdc42 [8]. Sy et al. showed that ZYX is frequently overexpressed in hepatocellular carcinoma and renders tumor cells with an invasive phenotype [9]. Recently, Zhong et al. found that decreased level of ZYX impairs invasive ability of colorectal cancer cells probably via the focal adhesion pathway [10]. However, the role of ZYX in invasion of GBM remains to be elucidated. Therefore, in this study, we explored the role of ZYX in the invasion of GBM as well as the possible mechanisms underlying the regulation of ZYX on GBM behaviors.
Materials and methods
Cell culture
Primary GBM cell (GBM1) were isolated from southwest hospital [11] and GBM cell lines LN229, T98G, and U87 were obtained ATCC. Human normal astrocytes HEB were obtained from the Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences (Guangzhou, China) [12]. All cell lines were characterized as mycoplasma negative by Myco-Lumi™ Luminescent Mycoplasma Detection Kit (Beyotime, Shanghai, China) and validated by STR DNA fingerprinting using the AmpFLSTR Identifiler kit (ThermoFisher Scientific, Waltham, MA, USA) according to manufacturer’s instructions semiannually. The STR profiles were compared with known ATCC fingerprints (www.ATCC.org) and with the Cell Line Integrated Molecular Authentication database version 0.1.200808 (http://bioinformatics.istge.it/clima/) [13]. The STR profiles matched known DNA fingerprints or were unique. The cells were cultured at 37 °C in a humidified incubator with 5% CO2 and 95% O2 in DMEM and supplemented with 10% fetal bovine serum (Gibco, Waltham, MA, USA), Plasmocin™ Prophylactic (InvivoGene, CA, USA), and penicillin:streptomycin (1:1000) (Life Technologies, Waltham, MA, USA).
Patients and tissues
The glioma samples were randomly collected from patients with glioma who underwent curative resection from Southwest Hospital (Chongqing, China) during the period of January 2014 and June 2016 glioma diagnosis was based on the World Health Organization (WHO) Classification of Central Nervous System Tumor (2016). This study was approved by the Ethics Committee of the Southwest Hospital. Cohorts of patient specimens from TCGA_Glioma database, Rembrandt database, and Grevendeel database (http://gliovis.bioinfo.cnio.es/) were reviewed to validate the prognostic value and function of ZYX in glioma. The median expression was used as the final cutoff value.
Immunohistochemistry and scoring
Immunohistochemical (IHC) staining was performed on tissue slides using an EnVision Kit (DAKO). After deparaffinized, rehydrated in graded ethanol, antigen retrieval and blocking, slides were incubated with anti-ZYX (1:500, ab109316, abcam) and anti-STMN1 (1:200, ab52630, abcam) at 4 °C for overnight. After washing with PBS, a horseradish peroxidase-conjugated secondary antibody (DAKO) was added and incubated at 37 °C for 30 min. Sections were stained by DAB (DAKO) and counterstained with hematoxylin. IHC scoring method was performed as previously described [14]. Briefly, to score the IHC staining data, five images of each sample were taken and the average integrated optical density was measured by Image Pro Plus 6.0 software.
Lentiviral infection procedures
To obtain ZYX overexpressing cells, the GBM cells (LN229 and GBM1) were transfected with the pHBLV-CMV-MCS-3flag-EF1-puro lentiviral vector carrying ZYX (Hanbo, Shanghai China). To obtain STMN1 overexpressing cells, the GBM cells (LN229 and GBM1) were transfected with the pGC-FU-CMV-EGFP lentiviral vector carrying STMN1 (Genechem, Shanghai China). Cells transfected with the empty vector (Mock) were identified as negative control. To generate stable ZYX knockdown cells, GBM cell (LN229 and GBM1) was transfected with lentiviral vector (Genechem) carrying self-complementary hairpin DNA fragments that could generate ZYX-specific shRNA and scrambled RNA as control. The shRNA sequences were list as below:
shZYX-1: 5′-TCCACATGAAGTGTTACAA-3′;
shZYX-2: 5′-GTTCCAAGTCCAGTACCAA-3′;
Scrambled: 5′-TTCTCCGAACGTGTCACGT-3′.
In addition, fresh culture medium with 6 μg/ml of puromycin was used to select stable cell lines.
Western blotting
Western blotting was conducted as previously described [15]. The commercial antibodies against ZYX and STMN1 were obtained from abcam and β-actin was purchased from Cell Signaling Technology.
Immunofluorescence staining
Immunofluorescence staining was conducted as previously described [16]. Primary antibodies for ZYX and STMN1 were obtained from abcam. Secondary antibodies were purchased from Invitrogen.
qRT-PCR
qRT-PCR was performed as previously described [17]. PCR primers were listed as below:
ACTB-F: 5′-CATGTACGTTGCTATCCAGGC-3′;
ACTB-R: 5′-CTCCTTAATGTCACGCACGAT-3′;
STMN1-F: 5′-TCAGCCCTCGGTCAAAAGAAT-3′;
STMN1-R: 5′-TTCTCGTGCTCTCGTTTCTCA-3′;
ZYX-F: 5′-TCTCCCGCGATCTCCGTTT-3′;
ZYX-R: 5′-CCGGAAGGGATTCACTTTGGG-3′.
Cell migration assay and cell invasion assay
LN229 and GBM1 cell suspension (3.0 × 104 cells/ml) was prepared in serum-free DMEM and 200 μl was inoculated into the upper chamber, while 700 μl complete medium was inoculated into the lower chamber. After continuous culture for 24 h, the chambers were fixed in 4% paraformaldehyde for 15 min followed by drying and staining with 0.5% crystal violet for 15 min at room temperature. For invasion assay, BD Matrigel™ was added into transwell chambers before the cells were seeded, the time of culturing was 48 h.
Animal xenograft
LN229 cells (2 × 105) and GBM1 cells (2 × 105) with pLVX-eGFP-luciferase lentivirus transfected with shCtrl RNA or shZYX were injected intracranially into the right brain of 5-week-old female NOD-SCID mice. The size of xenografts was evaluated with bioluminescence imaging (PerkinElmer, Waltham, Massachusetts, USA). Tumor-bearing mice were sacrificed when the animals became moribund. The tumors were collected for Harris hematoxylin and alcoholic eosin staining and immunohistochemistry.
RNA sequence and iTRAQ
RNA sequence and iTRAQ (Supplementary Tables 1 and 2) was conducted by Shanghai Oebiotech Co. Ltd. (Shanghai, China). Genes were defined as differentially expressed when logarithmic expression ratios were more than two-fold (P < 0.05). Proteins were defined as differentially expressed when logarithmic expression ratios were more than 1.5-fold (*P < 0.05).
GSEA analysis of differential genes
Download the TCGA, Rembrandt, and Gravendeel database gene and divide the database patients into high/low ZYX groups with the median of ZYX gene as the critical point, then output the matrix graph. Set relevant parameters according to the instructions of the web page, and output normalized enrichment score and nominal P value after GSEA analysis.
Statistical analysis
All experiments were performed at least three times. Results are presented as the mean ± SD by Student's t test using SPSS 20.0 software (IBM) and GraphPad Prism 5.0.
Correlation of ZYX and STMN1 expression was calculated using Pearson analysis. Survival curves were generated by Kaplan–Meier analysis. Survival curves were compared between groups using the log-rank test. All significant statistical differences were defined as *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
ZYX expression was increased in GBM cells
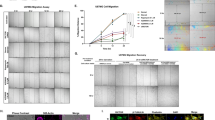
To investigate possible candidate genes responsible for cell migration and invasion in GBM, we first analyzed three invasion-related genesets, KEGG FOCAL ADHESION, GO CELL SUBSTRATE ADHESION, and GO CELL SUBSTRATE JUNCTION. Genes in the three genesets play critical roles in tumor invasion through regulation on interactions between cells and the extracellular matrix. A Venn diagram revealed that 26 genes were found in all three genesets (Fig. 1a). To further evaluate the potential involvement of the 26 genes in GBM invasion, we analyzed their expression levels in two widely used databases for GBM, TCGA_GBM and Rembrandt. Interestingly, the result showed that ZYX was the most highly increased gene in both databases (Fig. 1b), which implies a tight relationship between ZYX and mobility of GBM cells. Then, we examined the expression of ZYX proteins in a panel of GBM cell lines with a normal glia line as control. The data show that ZYX protein levels in GBM cell lines were higher than that in control cells (Fig. 1c). Furthermore, we also collected six freshly resected tumor tissues and their adjacent nontumor tissues. Consistent with the cell line results, ZYX protein levels in tumor tissues were higher than that in paired nontumor tissues (Fig. 1d). Therefore, ZYX appears to be a GBM-related gene and may be a candidate for invasion markers.
a The Venn diagram reveals that 26 genes overlap in three invasion-related genesets: KEGG FOCAL ADHESION, GO CELL SUBSTRATE ADHESION, and GO CELL SUBSTRATE JUNCTION. b The expression of the 26 overlapping genes from two public glioma databases, i.e., Rembrandt and TCGA_GBM. c ZYX protein expression is examined in one normal human normal glia cell line (HEB), three GBM cell lines (LN229, T98G, U87), and three primary GBM cells (GBM1, GBM2, GBM3) by Western blotting. Actin is used as a loading control. d ZYX protein expression is examined in six pairs GBM tumor tissues (T) and corresponding adjacent normal tissues (N) through Western blotting. Actin is used as loading control.
Increased ZYX was correlated with disease progression and poor prognosis of glioma
To comprehensively explore the pathological relevance of ZYX in gliomas, we analyzed ZYX expression in clinical glioma samples containing grade II, III, and IV or GBM. In our 84-case glioma cohort, immunohistochemistry staining revealed clear cytoplasmic localization of ZYX, and ZYX protein in GBM was much higher than that in grade II and III gliomas, but there was no difference in ZYX expression between grade II and grade III gliomas (Fig. 2a, b). In 12 cases of fresh human glioma samples, we also observed higher ZYX protein level in GBM than grade II and III gliomas through Western blotting (Fig. 2c). Analyses of three public glioma databases, i.e., TCGA_GBMLGG, Rembrandt, and Grevendeel, consistently revealed that the ZYX mRNA levels in GBM were significantly higher than that in grade II and III gliomas (Fig. 2d–f). Accordingly, ZYX was positively correlated with glioma progression. Moreover, Kaplan–Meier survival analysis on the three databases suggested that the patients with high ZYX levels showed shorter overall survival (OS) than those with low ZYX level in all gliomas as well as GBM only (Fig. 2g–i), which further supports the oncogenic role of ZYX in gliomas. Therefore, ZYX may act as a marker for disease progression and poor prognosis of glioma patients.
a Upper: Representative immunohistochemistry (IHC) image of glioma samples in different grades; Lower: Magnification of black dotted area. Scale bar = 100 μm (upper three images) or 20 μm (lower three images). b IHC score of ZYX in 84 glioma tissues of different grades. Data are expressed as the mean ± SD, **P < 0.01. c The expression of ZYX protein is examined in fresh glioma tissues of different grades through Western blotting. d The mRNA of ZYX in glioma with various grades in TCGA_Glioma database. e The mRNA of ZYX in glioma with various grades in Rembrandt database. f The mRNA of ZYX in glioma with various grades in Gravendeel database g Kaplan–Meier survival analysis of ZYX in glioma patients from TCGA_Glioma and TCGA_GBM databases. h Kaplan–Meier survival analysis of ZYX in glioma patients from Rembrandt and Rembrandt_GBM databases. i Kaplan–Meier survival analysis of ZYX in glioma patients from Gravendeel and Gravendeel_GBM databases.
High ZYX expression was related with enhanced invasiveness of glioma cells
Since ZYX plays pivotal roles in cell–extracellular substrate interactions, we speculated that ZYX might lead to glioma progression through promotion of invasion of tumor cells. To examine this hypothesis, we stably knocked down ZYX in LN229 cells (LN229/shZYX) and used scrambled shRNA as control (LN229/Ctrl) (Fig. 3a, b). LN229/shZYX and LN229/Ctrl cells were subjected to RNA-seq in triplicate. The data show that ZYX loss resulted in upregulation of 489 genes and downregulation of 219 genes (P < 0.05 and Fold ≥ 2) (Fig. 3c, left panel). Gene Ontology (GO) analysis revealed that the genes significantly altered by ZYX knockdown were enriched in several cell migration and invasion-related gene clusters under the context of biological process, such as cell adhesion, integrin-mediated signaling pathway, extracellular matrix organization, cell–matrix adhesion (Fig. 3c, right panel). Gene set enrichment assay (GSEA) further showed that ZYX level was positively correlated with the levels of genes responsible for glioma invasion (Fig. 3d), which were defined in our previous work [18]. The public databases were also analyzed and the results consistently indicated that high expression of ZYX enriched genes involved in glioma invasion and cell migration (Wu_Cell_Migration) [19] (Fig. 3e–g). Thus, both our own data and public databases identified potential involvement of ZYX in glioma invasion.
a ZYX mRNA expression in LN229/Ctrl, LN229/shZYX #1, and LN229/shZYX #2 cells using qRT-PCR. b ZYX protein expression in LN229/Ctrl, LN229/shZYX #1, and LN229/shZYX #2 cells using Western blotting. c Gene set enrichment analysis (GSEA) of LN229/Ctrl versus LN229/shZYX #1 in the context of Glioma_ Invasion genesets. d Heatmap (left panel) and gene ontology (GO) analysis (right panel) of differential genes through LN229/Ctrl versus LN229/shZYX #1 from RNA-Seq. e GSEA of different genes under ZYX high versus ZYX low in TCGA_GBM database. f GSEA of different genes under ZYX high versus ZYX low in Rembrandt database. g GSEA of different genes under ZYX high versus ZYX low in Gravendeel database.
ZYX knockdown prevents migration and invasion of GBM cells in vivo and in vitro
Next, we explored whether ZYX functionally affected the migration and invasion of GBM cells in Transwell assays. ZYX knockdown significantly inhibited serum-induced migration and invasion of tumor cells (Fig. 4a, b). We also intracranially inoculated control GBM cells or shZYX-transfected GBM cells into NOD-SCID mice followed by pathological analysis. We noticed that the margins of tumors formed by shZYX-transfected GBM cells was smooth and clearly separated tumor and nontumor areas (Fig. 4c). However, the margins of tumor formed by control GBM cells had obvious protrusions into nontumor areas (Fig. 4c). IHC staining of ZYX indicated that it was highly expressed in the invasive front of tumors formed by LN229/Ctrl cells (Fig. 4c). Consistent with the attenuated invasiveness of shZYX-transfected GBM cells, the mice with low ZYX GBM showed significantly longer survival time than those with control GBM (Fig. 4d, e). Therefore, ZYX was indeed positively correlated with invasiveness of GBM cells.
a The representative migration images (left panel) and statistical results (right panel) for LN229 and GBM1 cells with Ctrl, shZYX #1, and shZYX #2 transfection in a Transwell assay. Data are expressed as the mean ± SD, ***P < 0.001. Scale bar = 50 μm. b The representative invasion images (left panel) and statistical result of invaded cells (right panel) for LN229 and GBM1 cells with Ctrl, shZYX #1, and shZYX #2 transfection in a Transwell assay. Data are expressed as the mean ± SD, ***P < 0.001. Scale bar = 50 μm. c Representative H&E staining (left two images) and IHC images of ZYX (middle two images and right two images) in tumor area and adjacent nontumor area from xenografts formed by LN229/Ctrl and LN229/shZYX cells, respectively. Scale bar = 100 μm (left two and middles two images) or 25 μm (right two images). d Representative IHC images of ZYX in the core area of tumor and the invasive margin of xenograft implanted with GBM-1 cells. e Kaplan–Meier survival analysis on mice with GBM formed by LN229/Ctrl or LN229/shZYX #1 cells. OS: Overall survival. f Kaplan–Meier survival analysis on mice with GBM formed by GBM1/Ctrl or GBM1/shZYX #1 cells. OS: Overall survival.
ZYX regulated the transcription of STMN1 in GBM
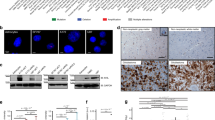
To explore the underlying mechanism of ZYX-mediated invasion, we performed iTRAQ-based Mass-Spectrum using LN229/Ctrl and LN229/shZYX cells to profile altered proteins related to ZYX loss. By comparing these results with the data from RNAseq, we found that STMN1, IGTA4, SRM, and ASNS were downregulated by ZYX knockdown in both RNA-seq data and mass-spectrum results (Fig. 5a). Among the four genes, we saw that STMN1 is known to play critical roles in tumor cell invasion [20,21,22,23,24,25,26,27]. However, ITGA4, SRM, and ASNS had not yet been linked to GBM. Hence, to identify reliable biomarkers for invasiveness of GBM, we further studied STMN1 in GBM cells. Consistent with the -omics data, both qRT-PCR and Western blotting confirmed the downregulation of STMN1 with ZYX knockdown in LN229 and GBM1 cells (Fig. 5b, c). In addition, immunofluorescence confirmed that lower expression of ZYX and STMN1 was detected in shZYX-transfected GBM cells compared with control GBM cells (Fig. 5d). Furthermore, we analyzed protein expression of ZYX and STMN1 through IHC (Fig. 5e) in a glioma cohort containing 61 cases (Supplementary Table 3). The Pearson correlation analysis showed that the protein expression of ZYX was positively correlated with that of STMN1 in the glioma cohort (Fig. 5f). GBM is classified into four subtypes, proneural, neural, mesenchymal, and classical, according to features of transcriptomes [28]. To further evaluate the relationship between ZYX and STMN1, we analyzed the mRNA expression of the two genes in GBM subtypes. Highest levels of ZYX mRNA was observed in classical subtype of GBM (Fig. 5g), but STMN1 mRNA expressed at the highest levels in the proneural subtype (Fig. 5h). Although the distribution of ZYX and STMN1 seemed inconsistent, correlation analysis in individual subtype revealed that ZYX and STMN1 were significantly positively correlated in proneural and classical subtypes (Fig. 5i, j). In the other two subtypes, however, no correlation was observed (Fig. 5k, l). Thus, the regulation of STMN1 by ZYX might be dependent on genetic background. Together, our results for the first time revealed that STMN1 may be a target of ZYX in GBM cells under specific genetic backgrounds.
a STMN1, IGTA4, SRM, and ASNS were consistently downregulated by ZYX knockdown in both RNA-seq and iTRAQ Mass-Spectrum using LN229/Ctrl and LN229/shZYX #1 cells. b The mRNA levels of STMN1 and ZYX in shCtrl and shZYX #1 GBM cells (LN229 and GBM1) by qRT-PCR. c The protein levels of STMN1 and ZYX in shCtrl and shZYX #1 GBM cells (LN229 and GBM1) by Western blotting. d Immunofluorescence staining showing the expression of STMN1 and ZYX in shCtrl and shZYX #1 GBM cells (LN229 and GBM1). Data are expressed as the mean ± SD, ***P < 0.001. Scale bar = 10 μm. e Representative IHC images of positive staining (upper two images) and negative staining (lower two images) for ZYX and STMN1, respectively, on continuous sections of human glioma tissues. Scale bar = 100 μm. f Pearson correlation of ZYX IHC scores and STMN1 IHC scores in 61 glioma tissues. g ZYX expression in different subtypes of GBM using TCGA_GBM database. Data are expressed as the mean ± SD. h STMN1 expression in different subtypes of GBM using TCGA_GBM database. Data are expressed as the mean ± SD. i–l Pearson correlations of ZYX mRNA and STMN1 mRNA in different subtypes of GBM using TCGA_GBM database.
STMN1 may function as a downstream target of ZYX in GBM cells
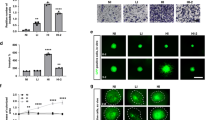
Because ZYX might regulate the expression of STMN1 in GBM, we profiled STMN1 expression in panel of GBM cells. Like ZYX, STMN1 was found to highly express in GBM cells compared with a normal astrocyte cell line, HEB (Fig. 6a). Moreover, we observed that the distribution of STMN1 in xenografts derived from LN229/Ctrl cells or LN229/shZYX cells in mice was consistent with that of ZYX (Fig. 6b versus Fig. 4c). In xenografts derived from LN229/Ctrl cells, the expression of STMN1 was obvious at the margin of tumor (Fig. 6b). To examine whether the overexpression of STMN1 could rescue the inhibition of tumor invasion due to ZYX knockdown, STMN1 was stably transfected into LN229 and GBM1 cells with or without ZYX knockdown (Fig. 6c). Migration assays using Transwell plates showed that STMN1 overexpression not only increased migration of GBM cells with ZYX but also partially rescued the inhibitory effect of ZYX knockdown on migration (Fig. 6d). Invasion assay also supported that STMN1 promoted invasion and rescued ZYX knockdown-induced invasion repression (Fig. 6e). Therefore, our results indicate that STMN1 could be a downstream mediator for ZYX functions in GBM cells.
a Western blotting shows the expression of STMN1 in GBM cell lines. b Representative IHC images of STMN1 in the tumor area and adjacent nontumor area from xenograft of LN229/Ctrl or LN229/shZYX cells. Scale bar = 100 μm (left two images) or 25 μm (right two images). c Western blotting shows the expression of ZYX and STMN1 in GBM cells (LN229 and GBM1) treated with Control shRNA (Ctrl), shZYX, Ctrl, or shZYX with STMN1 expression plasmid (STMN1). d Representative image and quantitative result for migration assay of GBM cells transfected with indicated plasmids. Scale bar = 50 μm. e Representative image and quantitative result for invasion assay of GBM cells transfected with indicated plasmids. Scale bar = 50 μm.
Discussion
In this study, our own data as well as public glioma databases consistently show that the expression of ZYX increased with the progression of glioma, and the highest level of ZYX expression was observed in GBM. Survival analysis further supported the oncogenic features of ZYX in glioma, including GBM. Functional analysis showed that ZYX promoted migration and invasion of GBM cells, potentially through STMN1 signaling. Although ZYX has been found to promote invasion of colorectal cancer cells and gastric cancer cells, neither the downstream targets of ZYX nor clinical relevance of ZYX in GBM has been reported. Our current study for the first time revealed the participation of ZYX in migration and invasion of GBM through the regulation of STMN1 levels.
For GBM, poor prognosis is generally considered to be consequence of high invasiveness. Our work indicates that high expression of ZYX may promote invasion of GBM cells in vitro and in vivo, which we believe leads to the poor prognosis of patients with high ZYX expression. Invasive growth depends on the increased mobility of tumor cells and interactions of tumor cells with the surrounding extracellular matrix [29]. Focal adhesions are known to pay critical roles during invasive growth of many kinds of cancers [30]. ZYX is one of the core proteins in focal adhesion and widely distributed in various cell types [31]. Functionally, ZYX can promote formation of and stabilize focal adhesions through regulation of the actin cytoskeleton [32, 33]. ZYX mediates interactions of cells and matrix by regulating assembly of the actin cytoskeleton, which ultimately affects cell migration, proliferation, and death [34]. Interestingly, TRIP6, a ZYX-related protein, is highly expressed in glioma, and glioma patients with high TRIP6 expression have a poor prognosis [35], which may also be related to the function of ZYX. In addition, ZYX also acts as an oncogene in several types of cancers, such as breast cancer [36] and melanoma [37].
The GO analysis of our RNAseq data revealed that forced expression of ZYX led to significant enrichment of the “biological process” categories, including hemidesmosome assembly, cell adhesion, extracellular matrix organization, homophilic cell adhesion via plasma membrane adhesion molecule, and cell–matrix adhesion. These categories reflect the known functions of ZYX, indicating that our RNAseq data were reliable. Ontology analysis also showed the enrichment of some molecular function categories, which have not yet been linked with ZYX, such as fibronectin binding, heparin binding, intracellular calcium activated chloride channel activity, Ras guanyl-nucleotide exchange factor activity, and phosphorelay sensor kinase activity. These molecular function categories might highlight the potential mechanisms by which ZYX regulates cell behaviors. Thus, our work herein further confirms the involvement of ZYX in the invasive ability of GBM cells and reveals several potential downstream pathways influenced by ZYX.
Although ZYX may enter the nucleus to regulate the expression of various genes and functions in the manner of a transcriptional factor [38, 39], our results did not identify its nuclear localization, which might imply that ZYX regulated STMN1 expression through other mediators. In glioma, LRRC4 [40], miR-9 [41], Malat1 [42], miR-16-1 [43], and PDGFRA [44] have been found to regulate STMN1 expression and functions. Accordingly, the regulation of STMN1 might be complicated and the mechanism on the regulation of STMN1 by ZYX is worth pursuing further.
STMN1 belongs to the stathmin family [45] and acts as a microtubule destabilizer to regulate microtubules dynamics [46, 47]. STMN1 plays a pivotal role in cell division and proliferation by regulating microtubule dynamics in tumor cells [48, 49]. In gliomas, STMN1 promotes tumor cell proliferation [41, 42] and decreased expression of STMN1 suppressed neoangiogenesis of gliomas [50]. Moreover, STMN1 correlates with tumor invasiveness [25, 51,52,53,54,55]. In brain tumors, Liang et al. observed that interference of STMN1 limits the spread of malignant gliomas in brain [56], and Marie et al. confirmed that STMN1 expression was significantly increased in malignant diffusely infiltrative astrocytomas compared with pilocytic astrocytoma [57].
Our current results show that overexpression of STMN1 led to enhanced invasion of GBM cells and could at least partially rescue the invasion deficiency caused by ZYX-knockdown. Thus, it is possible that ZYX promotes invasive growth of GBM cells through STMN1. Previous studies have shown the importance of actin/microtubule dynamics in both cell front membrane protrusion and cell rear retraction, which are two key processes of cell invasion [58,59,60]. Further evidence of cross-communication and signaling between the microtubule and actin cytoskeleton involves the microtubule-interacting protein STMN1 [26, 48, 61]. Thereby, our results imply that ZYX and STMN1 might synergistically promote the invasion of human GBM.
Our study provides insights on ZYX functions and reveals that ZYX may play an important role in the invasion of GBM, possibly through regulating the expression of STMN1. Moreover, ZYX correlates with glioma progression and worse prognosis of patients. ZYX may therefore be a biomarker for diagnosis and a potential druggable target for treatment.
References
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109.
Markert J. Glioblastoma multiforme: introduction. Cancer J. 2003;9:148.
Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011-2015. Neuro-Oncology 2018;20:1–86.
Oike T, Suzuki Y, Sugawara K, Shirai K, Noda SE, Tamaki T, et al. Radiotherapy plus concomitant adjuvant temozolomide for glioblastoma: Japanese mono-institutional results. PloS ONE. 2013;8:e78943.
Ntantie E, Allen MJ, Fletcher J, Nkembo AT, Lamango NS, Ikpatt OF. Suppression of focal adhesion formation may account for the suppression of cell migration, invasion and growth of non-small cell lung cancer cells following treatment with polyisoprenylated cysteinyl amide inhibitors. Oncotarget. 2018;9:25781–95.
Wang Y, McNiven MA. Invasive matrix degradation at focal adhesions occurs via protease recruitment by a FAK-p130Cas complex. J Cell Biol. 2012;196:375–85.
Pauli BU, Schwartz DE, Thonar EJ, Kuettner KE. Tumor invasion and host extracellular matrix. Cancer Metastasis Rev. 1983;2:129–52.
Yamamura M, Noguchi K, Nakano Y, Segawa E, Zushi Y, Takaoka K, et al. Functional analysis of Zyxin in cell migration and invasive potential of oral squamous cell carcinoma cells. Int J Oncol. 2013;42:873–80.
Sy SM, Lai PB, Pang E, Wong NL, To KF, Johnson PJ, et al. Novel identification of zyxin upregulations in the motile phenotype of hepatocellular carcinoma. Mod Pathol. 2006;19:1108–16.
Zhong C, Yu J, Li D, Jiang K, Tang Y, Yang M, et al. Zyxin as a potential cancer prognostic marker promotes the proliferation and metastasis of colorectal cancer cells. J Cell Physiol. 2019. https://doi.org/10.1002/jcp.28236.
Lv D, Ma QH, Duan JJ, Wu HB, Zhao XL, Yu SC, et al. Optimized dissociation protocol for isolating human glioma stem cells from tumorspheres via fluorescence-activated cell sorting. Cancer Lett. 2016;377:105–15.
Shu M, Zheng X, Wu S, Lu H, Leng T, Zhu W, et al. Targeting oncogenic miR-335 inhibits growth and invasion of malignant astrocytoma cells. Mol Cancer. 2011;10:59.
Romano P, Manniello A, Aresu O, Armento M, Cesaro M, Parodi B. Cell Line Data Base: structure and recent improvements towards molecular authentication of human cell lines. Nucleic Acids Res. 2009;37:925–32.
Tan J, Liu M, Zhang JY, Yao YL, Wang YX, Lin Y, et al. Capillary morphogenesis protein 2 is a novel prognostic biomarker and plays oncogenic roles in glioma. J Pathol. 2018;245:160–71.
Li Y, He ZC, Zhang XN, Liu Q, Chen C, Zhu Z, et al. Stanniocalcin-1 augments stem-like traits of glioblastoma cells through binding and activating NOTCH1. Cancer Lett. 2018;416:66–74.
Chen Q, Weng HY, Tang XP, Lin Y, Yuan Y, Li Q, et al. ARL4C stabilized by AKT/mTOR pathway promotes the invasion of PTEN-deficient primary human glioblastoma. J Pathol. 2019;247:266–78.
Zou H, Chen Q, Zhang A, Wang S, Wu H, Yuan Y, et al. MPC1 deficiency accelerates lung adenocarcinoma progression through the STAT3 pathway. Cell Death Dis. 2019;10:148.
Cao MF, Chen L, Dang WQ, Zhang XC, Zhang X, Shi Y, et al. Hybrids by tumor-associated macrophages x glioblastoma cells entail nuclear reprogramming and glioblastoma invasion. Cancer Lett. 2019;442:445–52.
Wu Y, Siadaty MS, Berens ME, Hampton GM, Theodorescu D. Overlapping gene expression profiles of cell migration and tumor invasion in human bladder cancer identify metallothionein 1E and nicotinamide N-methyltransferase as novel regulators of cell migration. Oncogene. 2008;27:6679–89.
Shu F, Zou X, Tuo H, She S, Huang J, Ren H, et al. Stathmin gene silencing suppresses proliferation, migration and invasion of gastric cancer cells via AKT/sCLU and STAT3 signaling. Int J Oncol. 2019;54:1086–98.
Aronova A, Min IM, Crowley MJP, Panjwani SJ, Finnerty BM, Scognamiglio T, et al. STMN1 is overexpressed in adrenocortical carcinoma and promotes a more aggressive phenotype in vitro. Ann Surg Oncol. 2018;25:792–800.
Yurong L, Biaoxue R, Wei L, Zongjuan M, Hongyang S, Ping F, et al. Stathmin overexpression is associated with growth, invasion and metastasis of lung adenocarcinoma. Oncotarget. 2017;8:26000–12.
Ni PZ, He JZ, Wu ZY, Ji X, Chen LQ, Xu XE, et al. Overexpression of Stathmin 1 correlates with poor prognosis and promotes cell migration and proliferation in oesophageal squamous cell carcinoma. Oncol Rep. 2017;38:3608–18.
Chen Y, Zhang Q, Ding C, Zhang X, Qiu X, Zhang Z. Stathmin1 overexpression in hypopharyngeal squamous cell carcinoma: a new promoter in FaDu cell proliferation and migration. Int J Oncol. 2017;50:31–40.
He X, Liao Y, Lu W, Xu G, Tong H, Ke J, et al. Elevated STMN1 promotes tumor growth and invasion in endometrial carcinoma. Tumour Biol. 2016;37:9951–8.
Byrne FL, Yang L, Phillips PA, Hansford LM, Fletcher JI, Ormandy CJ, et al. RNAi-mediated stathmin suppression reduces lung metastasis in an orthotopic neuroblastoma mouse model. Oncogene. 2014;33:882–90.
Akhtar J, Wang Z, Yu C, Li CS, Shi YL, Liu HJ. STMN-1 is a potential marker of lymph node metastasis in distal esophageal adenocarcinomas and silencing its expression can reverse malignant phenotype of tumor cells. BMC Cancer. 2014;14:28.
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110.
Paw I, Carpenter RC, Watabe K, Debinski W, Lo HW. Mechanisms regulating glioma invasion. Cancer Lett. 2015;362:1–7.
Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–74.
Beckerle MC. Identification of a new protein localized at sites of cell-substrate adhesion. J. Cell Biol. 1986;103:1679–87.
Nix DA, Fradelizi J, Bockholt S, Menichi B, Louvard D, Friederich E, et al. Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J Biol Chem. 2001;276:34759–67.
Han J, Liu G, Profirovic J, Niu J, Voyno-Yasenetskaya T. Zyxin is involved in thrombin signaling via interaction with PAR-1 receptor. FASEB J. 2009;23:4193–206.
Hirata H, Tatsumi H, Sokabe M. Zyxin emerges as a key player in the mechanotransduction at cell adhesive structures. Commun Integr Biol. 2008;1:192–5.
Lin VT, Lin VY, Lai YJ, Chen CS, Liu K, Lin WC, et al. TRIP6 regulates p27 KIP1 to promote tumorigenesis. Mol Cell Biol. 2013;33:1394–409.
Ma B, Cheng H, Gao R, Mu C, Chen L, Wu S, et al. Zyxin-Siah2-Lats2 axis mediates cooperation between Hippo and TGF-beta signalling pathways. Nat Commun. 2016;7:11123.
van der Gaag EJ, Leccia MT, Dekker SK, Jalbert NL, Amodeo DM, Byers HR. Role of zyxin in differential cell spreading and proliferation of melanoma cells and melanocytes. J Investig Dermatol. 2002;118:246–54.
Degenhardt YY, Silverstein S. Interaction of zyxin, a focal adhesion protein, with the e6 protein from human papillomavirus type 6 results in its nuclear translocation. J Virol. 2001;75:11791–802.
Grunewald TG, Kammerer U, Schulze E, Schindler D, Honig A, Zimmer M, et al. Silencing of LASP-1 influences zyxin localization, inhibits proliferation and reduces migration in breast cancer cells. Exp Cell Res. 2006;312:974–82.
Wang R, Wang Z, Yang J, Liu X, Wang L, Guo X, et al. LRRC4 inhibits the proliferation of human glioma cells by modulating the expression of STMN1 and microtubule polymerization. J Cell Biochem. 2011;112:3621–9.
Song Y, Mu L, Han X, Li Q, Dong B, Li H, et al. MicroRNA-9 inhibits vasculogenic mimicry of glioma cell lines by suppressing Stathmin expression. J Neurooncol. 2013;115:381–90.
Fu Z, Luo W, Wang J, Peng T, Sun G, Shi J, et al. Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma. Biochem Biophys Res Commun. 2017;492:480–6.
Li X, Ling N, Bai Y, Dong W, Hui GZ, Liu D, et al. MiR-16-1 plays a role in reducing migration and invasion of glioma cells. Anat Rec. 2013;296:427–32.
Jun HJ, Appleman VA, Wu HJ, Rose CM, Pineda JJ, Yeo AT, et al. A PDGFRalpha-driven mouse model of glioblastoma reveals a stathmin1-mediated mechanism of sensitivity to vinblastine. Nat Commun. 2018;9:3116.
Machado-Neto JA, Saad ST, Traina F. Stathmin 1 in normal and malignant hematopoiesis. BMB Rep. 2014;47:660–5.
Honnappa S, Cutting B, Jahnke W, Seelig J, Steinmetz MO. Thermodynamics of the Op18/stathmin-tubulin interaction. J Biol Chem. 2003;278:38926–34.
Steinmetz MO. Structure and thermodynamics of the tubulin-stathmin interaction. J Struct Biol. 2007;158:137–47.
Nemunaitis J. Stathmin 1: a protein with many tasks. New biomarker and potential target in cancer. Exp Opin Ther Targets. 2012;16:631–4.
Powrozek T, Mlak R, Krawczyk P, Barton S, Biernacka B, Malecka-Massalska T, et al. Retrospective analysis of second-line chemotherapy outcomes with paclitaxel or docetaxel in correlation with STMN1 polymorphism in advanced non-small cell lung cancer patients. Clin Transl Oncol. 2016;18:33–9.
Dong B, Mu L, Qin X, Qiao W, Liu X, Yang L, et al. Stathmin expression in glioma-derived microvascular endothelial cells: a novel therapeutic target. Oncol Rep. 2012;27:714–8.
Biaoxue R, Hua L, Wenlong G, Shuanying Y. Overexpression of stathmin promotes metastasis and growth of malignant solid tumors: a systemic review and meta-analysis. Oncotarget. 2016;7:78994–9007.
Li X, Wang L, Li T, You B, Shan Y, Shi S, et al. STMN1 overexpression correlates with biological behavior in human cutaneous squamous cell carcinoma. Pathol Res Pract. 2015;211:816–23.
Nie W, Xu MD, Gan L, Huang H, Xiu Q, Li B. Overexpression of stathmin 1 is a poor prognostic biomarker in non-small cell lung cancer. Lab Investig. 2015;95:56–64.
Tan HT, Wu W, Ng YZ, Zhang X, Yan B, Ong CW, et al. Proteomic analysis of colorectal cancer metastasis: stathmin-1 revealed as a player in cancer cell migration and prognostic marker. J Proteome Res. 2012;11:1433–45.
Liu F, Sun YL, Xu Y, Liu F, Wang LS, Zhao XH. Expression and phosphorylation of stathmin correlate with cell migration in esophageal squamous cell carcinoma. Oncol Rep. 2013;29:419–24.
Liang XJ, Choi Y, Sackett DL, Park JK. Nitrosoureas inhibit the stathmin-mediated migration and invasion of malignant glioma cells. Cancer Res. 2008;68:5267–72.
Marie SK, Oba-Shinjo SM, da Silva R, Gimenez M, Nunes Reis G, Tassan JP, et al. Stathmin involvement in the maternal embryonic leucine zipper kinase pathway in glioblastoma. Proteome Sci. 2016;14:6.
Niethammer P, Bastiaens P, Karsenti E. Stathmin-tubulin interaction gradients in motile and mitotic cells. Science. 2004;303:1862–6.
Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, et al. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–94.
Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–9.
Baldassarre G, Belletti B, Nicoloso MS, Schiappacassi M, Vecchione A, Spessotto P, et al. p27(Kip1)-stathmin interaction influences sarcoma cell migration and invasion. Cancer Cell. 2005;7:51–63.
Acknowledgements
This study was supported by the grants from the National Key Research and Development Program (No. 2016YFA0101203) (X.-W.B.) and the National Natural Science Foundation of China (No. 81821003) (X.-W.B.).
Author information
Authors and Affiliations
Contributions
Conception and design: Xiu-Wu Bian, Yan Wang, Xia Zhang, You-Hong Cui. Development of methodology: Tao Luo, Xian-Mei Wen, Yi Jiang, Qian Chen, Kai-Di Yang, Li-Hong Wang, Ying Luo, Ye Yuan, Chun-Hua Luo, Xiang Zhang, Ze-Xuan Yan, Wen-Juan Fu, Yu-Huan Tan, Qin Niu, Jing-Fang Xiao, Lu Chen, Jiao Wang, Jiafeng Huang. Acquisition of data: Tao Luo, Xian-Mei Wen. Analysis and interpretation of data: Yan Wang. Writing and revision of the manuscript: Xiu-Wu Bian, Yan Wang.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wen, XM., Luo, T., Jiang, Y. et al. Zyxin (ZYX) promotes invasion and acts as a biomarker for aggressive phenotypes of human glioblastoma multiforme. Lab Invest 100, 812–823 (2020). https://doi.org/10.1038/s41374-019-0368-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-019-0368-9
This article is cited by
-
Nitroproteomics is instrumental for stratification and targeted treatments of astrocytoma patients: expert recommendations for advanced 3PM approach with improved individual outcomes
EPMA Journal (2023)
-
Angiocrine extracellular vesicles impose mesenchymal reprogramming upon proneural glioma stem cells
Nature Communications (2022)
-
Hyperbaric oxygen promotes not only glioblastoma proliferation but also chemosensitization by inhibiting HIF1α/HIF2α-Sox2
Cell Death Discovery (2021)
-
Cell density-dependent proteolysis by HtrA1 induces translocation of zyxin to the nucleus and increased cell survival
Cell Death & Disease (2020)