Abstract
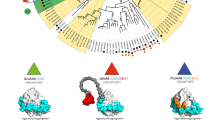
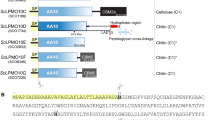
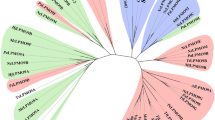
Lytic polysaccharide monooxygenases (LPMOs) are copper-containing enzymes that play a key role in the oxidative degradation of various biopolymers such as cellulose and chitin. While hunting for new LPMOs, we identified a new family of proteins, defined here as X325, in various fungal lineages. The three-dimensional structure of X325 revealed an overall LPMO fold and a His brace with an additional Asp ligand to Cu(II). Although LPMO-type activity of X325 members was initially expected, we demonstrated that X325 members do not perform oxidative cleavage of polysaccharides, establishing that X325s are not LPMOs. Investigations of the biological role of X325 in the ectomycorrhizal fungus Laccaria bicolor revealed exposure of the X325 protein at the interface between fungal hyphae and tree rootlet cells. Our results provide insights into a family of copper-containing proteins, which is widespread in the fungal kingdom and is evolutionarily related to LPMOs, but has diverged to biological functions other than polysaccharide degradation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
LaX325 nucleotide sequence was deposited in GenBank under accession number MK088083. The X-ray structures of LaX325 were deposited in the Protein Data Bank under accession numbers 6IBH, 6IBI and 6IBJ. Raw EPR data are available on request through the Research Data York (https://doi.org/10.15124/a034974e-2782-415e-8b02-2b6e4098760e).
References
Vaaje-Kolstad, G. et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222 (2010).
Quinlan, R. J. et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl Acad. Sci. USA 108, 15079–15084 (2011).
Horn, S. J., Vaaje-Kolstad, G., Westereng, B. & Eijsink, V. G. Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 5, 45 (2012).
Bissaro, B. et al. Oxidative cleavage of polysaccharides by monocopper enzymes depends on H2O2. Nat. Chem. Biol. 13, 1123 (2017).
Johansen, K. S. Discovery and industrial applications of lytic polysaccharide mono-oxygenases. Biochem. Soc. Trans. 44, 143–149 (2016).
Bennati-Granier, C. et al. Substrate specificity and regioselectivity of fungal AA9 lytic polysaccharide monooxygenases secreted by Podospora anserina. Biotechnol. Biofuels 8, 90 (2015).
Petrović, D. M. et al. Methylation of the N-terminal histidine protects a lytic polysaccharide monooxygenase from auto-oxidative inactivation. Protein Sci. 27, 1636–1650 (2018).
Ciano, L., Davies, G. J., Tolman, W. B. & Walton, P. H. Bracing copper for the catalytic oxidation of C–H bonds. Nat. Catal. 1, 571–577 (2018).
Gilbert, H. J., Knox, J. P. & Boraston, A. B. Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr. Opin. Struc. Biol. 23, 669–677 (2013).
Hemsworth, G. R., Henrissat, B., Davies, G. J. & Walton, P. H. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat. Chem. Biol. 10, 122–126 (2014).
Lo Leggio, L. et al. Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nat. Commun. 6, 5961 (2015).
Vu, V. V., Beeson, W. T., Span, E. A., Farquhar, E. R. & Marletta, M. A. A family of starch-active polysaccharide monooxygenases. Proc. Natl Acad. Sci. USA 111, 13822–13827 (2014).
Sabbadin, F. et al. An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion. Nat. Commun. 9, 756 (2018).
Couturier, M. et al. Lytic xylan oxidases from wood-decay fungi unlock biomass degradation. Nat. Chem. Biol. 14, 306 (2018).
Filiatrault-Chastel, C. et al. AA16, a new lytic polysaccharide monooxygenase family identified in fungal secretomes. Biotech. Biofuels 12, 55 (2019).
Berrin, J.-G., Rosso, M.-N. & Abou Hachem, M. Fungal secretomics to probe the biological functions of lytic polysaccharide monooxygenases. Carbohydr. Res. 448, 155–160 (2017).
Navarro, D. et al. Fast solubilization of recalcitrant cellulosic biomass by the basidiomycete fungus Laetisaria arvalis involves successive secretion of oxidative and hydrolytic enzymes. Biotechnol. Biofuels 7, 143 (2014).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Forsberg, Z. et al. Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases. Proc. Natl Acad. Sci. USA 111, 8446–8451 (2014).
Low, M. & Saltiel, A. Structural and functional roles of glycosyl-phosphatidylinositol in membranes. Science 239, 268–275 (1988).
Berbee, M. L., James, T. Y. & Strullu-Derrien, C. Early diverging fungi: diversity and impact at the dawn of terrestrial life. Annu. Rev. Microbiol. 71, 41–60 (2017).
Tandrup, T., Frandsen, K. E. H., Johansen, K. S., Berrin, J.-G. & Lo Leggio, L. Recent insights into lytic polysaccharide monooxygenases (LPMOs). Biochem. Soc. Trans. 46, 1431–1447 (2018).
Holm, L. & Laakso, L. M. Dali server update. Nucleic Acids Res. 44, W351–W355 (2016).
Vu, V. V. & Ngo, S. T. Copper active site in polysaccharide monooxygenases. Coord. Chem. Rev. 368, 134–157 (2018).
Chiu, E. et al. Structural basis for the enhancement of virulence by viral spindles and their in vivo crystallization. Proc. Natl Acad. Sci. USA 112, 3973–3978 (2015).
Tan, T.-C. et al. Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation. Nat. Commun. 6, 7542 (2015).
Lawton, T. J., Kenney, G. E., Hurley, J. D. & Rosenzweig, A. C. The CopC family: structural and bioinformatic insights into a diverse group of periplasmic copper binding proteins. Biochemistry 55, 2278–2290 (2016).
Ross, M. O. et al. Particulate methane monooxygenase contains only mononuclear copper centers. Science 364, 566–570 (2019).
Liew, E. F., Tong, D., Coleman, N. V. & Holmes, A. J. Mutagenesis of the hydrocarbon monooxygenase indicates a metal centre in subunit-C, and not subunit-B, is essential for copper-containing membrane monooxygenase activity. Microbiology 160, 1267–1277 (2014).
Cao, L., Caldararu, O., Rosenzweig, A. C. & Ryde, U. Quantum refinement does not support dinuclear copper sites in crystal structures of particulate methane monooxygenase. Angew. Chem. Int. Ed. Eng. 57, 162–166 (2018).
Peisach, J. & Blumberg, W. E. Structural implications derived from the analysis of electron paramagnetic resonance spectra of natural and artificial copper proteins. Arch. Biochem. Biophys. 165, 691–708 (1974).
Kuusk, S. et al. Kinetics of H2O2-driven degradation of chitin by a bacterial lytic polysaccharide monooxygenase. J. Biol. Chem. 293, 523–531 (2018).
Hangasky, J. A., Iavarone, A. T. & Marletta, M. A. Reactivity of O2 versus H2O2 with polysaccharide monooxygenases. Proc. Natl Acad. Sci. USA 115, 4915–4920 (2018).
Phillips, C. M., Beeson, W. T., Cate, J. H. & Marletta, M. A. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 6, 1399–1406 (2011).
Bey, M. et al. Cello-oligosaccharide oxidation reveals differences between two lytic polysaccharide monooxygenases (family GH61) from Podospora anserina. Appl. Environ. Microbiol. 79, 488–496 (2013).
Chen, K. H. et al. Bacteriohemerythrin bolsters the activity of the particulate methane monooxygenase (pMMO) in Methylococcus capsulatus (Bath). J. Inorg. Biochem. 111, 10–17 (2012).
Bernardes et al. Carbohydrate binding modules enhance cellulose enzymatic hydrolysis by increasing access of cellulases to the substrate. Carbohydr. Polym. 211, 57–68 (2019).
Shah, F. et al. Ectomycorrhizal fungi decompose soil organic matter using oxidative mechanisms adapted from saprotrophic ancestors. New Phytol. 209, 1705–1719 (2016).
Balestrini R. M. & Kottke I. In Molecular Mycorrhizal Symbiosis (ed. Martin, F.) 47–62 (John Wiley & Sons, 2016).
Balestrini, R., Hahn, M. G., Faccio, A., Mendgen, K. & Bonfante, P. Differential localization of carbohydrate epitopes in plant cell walls in the presence and absence of arbuscular mycorrhizal fungi. Plant Physiol. 111, 203–213 (1996).
Balestrini, R. & Bonfante, P. Cell wall remodeling in mycorrhizal symbiosis: a way towards biotrophism. Front. Plant Sci. 5, 237 (2014).
Yamazaki, H., Tanaka, A., Kaneko, J.-i, Ohta, A. & Horiuchi, H. Aspergillus nidulans ChiA is a glycosylphosphatidylinositol (GPI)-anchored chitinase specifically localized at polarized growth sites. Fungal Genet. Biol. 45, 963–972 (2008).
Nakajima, M., Yamashita, T., Takahashi, M., Nakano, Y. & Takeda, T. A novel glycosylphosphatidylinositol-anchored glycoside hydrolase from Ustilago esculenta functions in β-1,3-glucan degradation. Appl. Environ. Microbiol. 78, 5682–5689 (2012).
Smith, H. W. & Marshall, C. J. Regulation of cell signalling by uPAR. Nat. Rev. Mol. Cell Biol. 11, 23–36 (2010).
Gaggelli, E., Kozlowski, H., Valensin, D. & Valensin, G. Copper homeostasis and neurodegenerative disorders (Alzheimer’s, prion, and Parkinson’s diseases and amyotrophic lateral sclerosis). Chem. Rev. 106, 1995–2044 (2006).
García-Santamarina, S. & Thiele, D. J. Copper at the fungal pathogen–host axis. J. Biol. Chem. 290, 18945–18953 (2015).
Bolchi, A. et al. Genome-wide inventory of metal homeostasis-related gene products including a functional phytochelatin synthase in the hypogeous mycorrhizal fungus Tuber melanosporum. Fungal Genet. Biol. 48, 573–584 (2011).
Lalucque, H. et al. IDC2 and IDC3, two genes involved in cell non-autonomous signaling of fruiting body development in the model fungus Podospora anserina. Dev. Biol. 421, 126–138 (2017).
Green, K. A. et al. SymB and SymC, two membrane associated proteins, are required for Epichloë festucae hyphal cell–cell fusion and maintenance of a mutualistic interaction with Lolium perenne. Mol. Microbiol. 103, 657–677 (2016).
García-Santamarina, S. et al. A lytic polysaccharide monooxygenase-like protein functions in copper import and fungal meningitis. Nat. Chem. Biol. https://doi.org/10.1038/s41589-019-0437-9.
Kohler, A. et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat. Genet. 47, 410–415 (2015).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Lemoine, F. et al. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature 556, 452–456 (2018).
Huson, D. H. & Scornavacca, C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61, 1061–1067 (2012).
Haon, M. et al. Recombinant protein production facility for fungal biomass-degrading enzymes using the yeast Pichia pastoris. Front. Microbiol. 6, 1002 (2015).
Pitarch, A., Sánchez, M., Nombela, C. & Gil, C. Sequential fractionation and two-dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol. Cell. Proteom. 1, 967–982 (2002).
Felten, J. et al. The ectomycorrhizal fungus Laccaria bicolor stimulates lateral root formation in poplar and Arabidopsis through auxin transport and signaling. Plant Physiol. 151, 1991–2005 (2009).
Martin, F. et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 452, 88–92 (2008).
Gorrec, F. The MORPHEUS protein crystallization screen. J. Appl. Crystallogr. 42, 1035–1042 (2009).
Terwilliger, T. C. et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Cryst. D Biol. Crystallogr. 65, 582–601 (2009).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 (2010).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Zeldin, O. B., Gerstel, M. & Garman, E. F. RADDOSE-3D: time- and space-resolved modelling of dose in macromolecular crystallography. J. Appl. Crystallogr. 46, 1225–1230 (2013).
Acknowledgements
A.L. was funded by a Marie Curie Individual Fellowship within the Horizon 2020 Research and Innovation Framework Programme (748758). The Danish Ministry of Higher Education and Science through the Instrument Center DANSCATT funded travel to synchrotrons. We would also like to acknowledge MAX-IV, Lund, Sweden, the ESRF, Grenoble, France and DESY, Hamburg, Germany for synchrotron beamtime and related assistance. We thank J.-C. Poulsen for technical assistance; A. Kohler and D. Thiele for helpful discussions; F. Chaspoul for assistance with ICP-MS analyses and R. Balestrini (Institute for Sustainable Plant Protection, Italy) for providing gold-labeled WGA lectin. L.L.L. and T.T. thank the Novo Nordisk Foundation for funding through grant NF17SA0027704. K.E.H.F. thanks the Carlsberg Foundation through an Internationalization Postdoc Fellowship (grants CF16-0673 and CF17-0533) and the EU, framework of the Marie Curie FP7 COFUND People Programme (AgreenSkills+ fellowship 609398) for financial support. L.L.L., T.T. and K.E.H.F. are members of ISBUC Integrative Structural Biology at the University of Copenhagen (https://isbuc.ku.dk/). K.S.J. thanks the Novo Nordisk Foundation for funding through grant NNF17SA0027704. P.H.W. and L.C. thank the UK Biotechnology and Biological Sciences Research Council (BB/L021633/1) for funding. F.M. is funded by the French National Research Agency through the Laboratory of Excellence Advanced Research on the Biology of Tree and Forest Ecosystems (grant ANR-11-LABX 0002 01). A.Z. thanks the French Embassy in Greece, together with the French Ministry of Higher Education, Research and Innovation, for the scholarship ‘Séjour scientifique de haut niveau’ (SSHN). The electron microscopy experiments were performed on the PiCSL-FBI core facility (IBDM, Marseille), member of the France-BioImaging national research infrastructure (ANR-10-INBS-04).
Author information
Authors and Affiliations
Contributions
A.L., D.N. and M.-N.R. identified the new proteins. A.L. and B.H. performed bioinformatic analyses. A.L., S.G. and M.H. performed recombinant protein production and purification. T.T. and K.E.H.F. crystallized LaX325, determined and analyzed the X-ray crystal structure. T.T. collected X-ray data. K.E.H.F. made relevant figures and tables. L.L.L. directed the crystallographic studies. K.E.H.F., K.S.J. and L.L.L. analyzed the structure. K.E.H.F. and L.L.L. drafted relevant parts of the manuscript. A.L., B.B. and A.Z. performed enzyme assays. M.F. and D.R. performed mass spectrometry analyses. L.C. and P.H.W. conceived and carried out the EPR study. A.L. and F.Z. performed confocal microscopy under the supervision of F.M. A.L. and N.B. performed transmission electron microscopy. J.-G.B. coordinated the work. A.L. and J.-G.B. organized the data and drafted the manuscript. All authors made comments on the manuscript and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–6 and Supplementary Figs. 1–9.
Rights and permissions
About this article
Cite this article
Labourel, A., Frandsen, K.E.H., Zhang, F. et al. A fungal family of lytic polysaccharide monooxygenase-like copper proteins. Nat Chem Biol 16, 345–350 (2020). https://doi.org/10.1038/s41589-019-0438-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0438-8
This article is cited by
-
A seven-transmembrane methyltransferase catalysing N-terminal histidine methylation of lytic polysaccharide monooxygenases
Nature Communications (2023)
-
Evaluation of endoglucanase and xylanase production by Aspergillus tamarii cultivated in agro-industrial lignocellulosic biomasses
Folia Microbiologica (2022)
-
Orchestrating copper binding: structure and variations on the cupredoxin fold
JBIC Journal of Biological Inorganic Chemistry (2022)
-
Recent Computational Insights into the Oxygen Activation by Copper-Dependent Metalloenzymes
Topics in Catalysis (2022)
-
Novel molecular biological tools for the efficient expression of fungal lytic polysaccharide monooxygenases in Pichia pastoris
Biotechnology for Biofuels (2021)