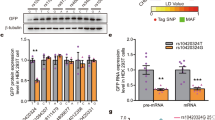
Abstract
High impulsive and aggressive traits associate with poor behavioural self-control. Despite their importance in predicting behavioural negative outcomes including suicide, the molecular mechanisms underlying the expression of impulsive and aggressive traits remain poorly understood. Here, we identified and characterized a novel long noncoding RNA (lncRNA), acting as a regulator of the monoamine oxidase A (MAOA) gene in the brain, and named it MAOA-associated lncRNA (MAALIN). Our results show that in the brain of suicide completers, MAALIN is regulated by a combination of epigenetic mechanisms including DNA methylation and chromatin modifications. Elevated MAALIN in the dentate gyrus of impulsive-aggressive suicides was associated with lower MAOA expression. Viral overexpression of MAALIN in neuroprogenitor cells decreased MAOA expression while CRISPR-mediated knock out resulted in elevated MAOA expression. Using viral-mediated gene transfer, we confirmed that MAALIN in the hippocampus significantly decreases MAOA expression and exacerbates the expression of impulsive-aggressive behavioural traits in CD1 aggressive mice. Overall, our findings suggest that variations in DNA methylation mediate the differential expression of a novel lncRNA that acts on MAOA expression to regulate impulsive-aggressive behaviours.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Contini V, Marques FZ, Garcia CE, Hutz MH, Bau CH. MAOA-uVNTR polymorphism in a Brazilian sample: further support for the association with impulsive behaviors and alcohol dependence. Am J Med Genet Part B, Neuropsychiatr Genet. 2006;141B:305–8.
Gallardo-Pujol D, Andres-Pueyo A, Maydeu-Olivares A. MAOA genotype, social exclusion and aggression: an experimental test of a gene-environment interaction. Genes, Brain, Behav. 2013;12:140–5.
Kuepper Y, Grant P, Wielpuetz C, Hennig J. MAOA-uVNTR genotype predicts interindividual differences in experimental aggressiveness as a function of the degree of provocation. Behav Brain Res. 2013;247:73–8.
McGirr A, Turecki G. The relationship of impulsive aggressiveness to suicidality and other depression-linked behaviors. Curr Psychiatry Rep. 2007;9:460–6.
Reti IM, Xu JZ, Yanofski J, McKibben J, Uhart M, Cheng YJ, et al. Monoamine oxidase A regulates antisocial personality in whites with no history of physical abuse. Compr Psychiatry. 2011;52:188–94.
Saito T, Lachman HM, Diaz L, Hallikainen T, Kauhanen J, Salonen JT, et al. Analysis of monoamine oxidase A (MAOA) promoter polymorphism in Finnish male alcoholics. Psychiatry Res. 2002;109:113–9.
Williams LM, Gatt JM, Kuan SA, Dobson-Stone C, Palmer DM, Paul RH, et al. A polymorphism of the MAOA gene is associated with emotional brain markers and personality traits on an antisocial index. Neuropsychopharmacology. 2009;34:1797–809.
Brezo J, Paris J, Hebert M, Vitaro F, Tremblay R, Turecki G. Broad and narrow personality traits as markers of one-time and repeated suicide attempts: a population-based study. BMC Psychiatry. 2008;8:15.
Brezo J, Paris J, Vitaro F, Hebert M, Tremblay RE, Turecki G. Predicting suicide attempts in young adults with histories of childhood abuse. Br J Psychiatry. 2008;193:134–9.
Turecki G, Brent DA. Suicide and suicidal behaviour. Lancet. 2016;387:1227–39.
Brent DA, Bridge J, Johnson BA, Connolly J. Suicidal behavior runs in families. A controlled family study of adolescent suicide victims. Arch Gen Psychiatry. 1996;53:1145–52.
McGirr A, Alda M, Seguin M, Cabot S, Lesage A, Turecki G. Familial aggregation of suicide explained by cluster B traits: a three-group family study of suicide controlling for major depressive disorder. Am J Psychiatry. 2009;166:1124–34.
McGowan PO, Suderman M, Sasaki A, Huang TC, Hallett M, Meaney MJ, et al. Broad epigenetic signature of maternal care in the brain of adult rats. PLoS One. 2011;6:e14739.
Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–54.
Zhang TY, Hellstrom IC, Bagot RC, Wen X, Diorio J, Meaney MJ. Maternal care and DNA methylation of a glutamic acid decarboxylase 1 promoter in rat hippocampus. J Neurosci. 2010;30:13130–7.
Labonte B, Yerko V, Gross J, Mechawar N, Meaney MJ, Szyf M, et al. Differential glucocorticoid receptor exon 1(B), 1(C), and 1(H) expression and methylation in suicide completers with a history of childhood abuse. Biol Psychiatry. 2012;72:41–8.
McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonte B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–8.
Labonte B, Suderman M, Maussion G, Lopez JP, Navaro L, Yerko V, et al. Genome-wide methylation changes in the brains of suicide completers. Am J Psychiatry. 2013;170:511–20. https://doi.org/10.1176/appi.ajp.2012.12050627.
Labonte B, Suderman M, Maussion G, Navaro L, Yerko V, Mahar I, et al. Genome-wide epigenetic regulation by early-life trauma. Arch Gen Psychiatry. 2012;69:722–31.
Brunner HG, Nelen M, Breakefield XO, Ropers HH, van Oost BA. Abnormal behavior associated with a point mutation in the structural gene for monoamine oxidase A. Science. 1993;262:578–80.
Cases O, Seif I, Grimsby J, Gaspar P, Chen K, Pournin S, et al. Aggressive behavior and altered amounts of brain serotonin and norepinephrine in mice lacking MAOA. Science. 1995;268:1763–6.
Scott AL, Bortolato M, Chen K, Shih JC. Novel monoamine oxidase A knock out mice with human-like spontaneous mutation. Neuroreport. 2008;19:739–43.
Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–7.
Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–66. https://doi.org/10.1038/nn.2436. Epub 8 Nov 2009.
Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–53.
Matevossian A, Akbarian S. Neuronal nuclei isolation from human postmortem brain tissue. J Vis Exp. 2008;20:pii: 914. https://doi.org/10.3791/914.
Coolen MW, Statham AL, Gardiner-Garden M, Clark SJ. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements. Nucleic Acids Res. 2007;35:e119.
Tursun B, Cochella L, Carrera I, Hobert O. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS One. 2009;4:e4625.
Golden SA, Covington HE 3rd, Berton O, Russo SJ. A standardized protocol for repeated social defeat stress in mice. Nat Protoc. 2011;6:1183–91.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36.
Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9.
Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–61.
R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2018.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Plaisier SB, Taschereau R, Wong JA, Graeber TG. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38:e169.
Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58.
Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–62.
Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–66.
Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7.
Bagot RC, Cates HM, Purushothaman I, Lorsch ZS, Walker DM, Wang J, et al. Circuit-wide transcriptional profiling reveals brain region-specific gene networks regulating depression susceptibility. Neuron. 2016;90:969–83.
Labonte B, Engmann O, Purushothaman I, Menard C, Wang J, Tan C, et al. Sex-specific transcriptional signatures in human depression. Nat Med. 2017;23:1102–11. https://doi.org/10.1038/nm.4386. Epub 21 Aug 2017.
Koolhaas JM, Coppens CM, de Boer SF, Buwalda B, Meerlo P, Timmermans PJ. The resident-intruder paradigm: a standardized test for aggression, violence and social stress. J Vis Exp. 2013;77:e4367.
Golden SA, Heshmati M, Flanigan M, Christoffel DJ, Guise K, Pfau ML, et al. Basal forebrain projections to the lateral habenula modulate aggression reward. Nature. 2016;534:688–92.
Huang X, Luo YL, Mao YS, Ji JL. The link between long noncoding RNAs and depression. Progress Neuro-Psychopharmacol Biol Psychiatry. 2017;73:73–8. https://doi.org/10.1016/j.pnpbp.2016.06.004. 16 Jun Epub 2016.
Roberts TC, Morris KV, Wood MJ. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos Trans R Soc Lond Ser B, Biol Sci. 2014;369:pii: 20130507. https://doi.org/10.1098/rstb.2013.0507.
Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16:324–37.
Dorfman HM, Meyer-Lindenberg A, Buckholtz JW. Neurobiological mechanisms for impulsive-aggression: the role of MAOA. Curr Topics Behav Neurosci. 2014;17:297–313. https://doi.org/10.1007/7854_2013_272.
Checknita D, Maussion G, Labonte B, Comai S, Tremblay RE, Vitaro F, et al. Monoamine oxidase A gene promoter methylation and transcriptional downregulation in an offender population with antisocial personality disorder. Br J Psychiatry. 2015;206:216–22.
Hinds HL, Hendriks RW, Craig IW, Chen ZY. Characterization of a highly polymorphic region near the first exon of the human MAOA gene containing a GT dinucleotide and a novel VNTR motif. Genomics. 1992;13:896–7.
Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206.
Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4.
Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149:819–31.
Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–81.
Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–3.
Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8.
Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51:349–59.
Zhou L, Sun K, Zhao Y, Zhang S, Wang X, Li Y, et al. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat Commun. 2015;6:10026.
Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6:7743.
O'Leary VB, Ovsepian SV, Carrascosa LG, Buske FA, Radulovic V, Niyazi M, et al. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep. 2015;11:474–85.
Kalwa M, Hanzelmann S, Otto S, Kuo CC, Franzen J, Joussen S, et al. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 2016;44:10631–43.
Deaton AM, Webb S, Kerr AR, Illingworth RS, Guy J, Andrews R, et al. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21:1074–86.
Iwamoto K, Bundo M, Ueda J, Oldham MC, Ukai W, Hashimoto E, et al. Neurons show distinctive DNA methylation profile and higher interindividual variations compared with non-neurons. Genome Res. 2011;21:688–96.
Ladd-Acosta C, Pevsner J, Sabunciyan S, Yolken RH, Webster MJ, Dinkins T, et al. DNA methylation signatures within the human brain. Am J Hum Genet. 2007;81:1304–15.
GBD 2015 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1545–602.
Lesch KP, Merschdorf U. Impulsivity, aggression, and serotonin: a molecular psychobiological perspective. Behav Sci Law. 2000;18:581–604.
Lewis AS, Pittenger ST, Mineur YS, Stout D, Smith PH, Picciotto MR. Bidirectional regulation of aggression in mice by hippocampal alpha-7 nicotinic acetylcholine receptors. Neuropsychopharmacology. 2018;43:1267–75.
Mitchell MR, Potenza MN. Recent insights into the neurobiology of impulsivity. Curr Addict Rep. 2014;1:309–19.
Caspi A, McClay J, Moffitt TE, Mill J, Martin J, Craig IW, et al. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–4.
Kim-Cohen J, Caspi A, Taylor A, Williams B, Newcombe R, Craig IW, et al. MAOA, maltreatment, and gene-environment interaction predicting children's mental health: new evidence and a meta-analysis. Mol Psychiatry. 2006;11:903–13.
Bortolato M, Chen K, Godar SC, Chen G, Wu W, Rebrin I, et al. Social deficits and perseverative behaviors, but not overt aggression, in MAO-A hypomorphic mice. Neuropsychopharmacology. 2011;36:2674–88.
Acknowledgements
GT holds a Canada Research Chair (Tier 1) and a NARSAD Distinguished Investigator Award. He is supported by grants from the Canadian Institute of Health Research (CIHR) (FDN148374 and EGM141899), by the FRQS through the Quebec Network on Suicide, Mood Disorders and Related Disorders. BL holds a Sentinelle Nord Research Chair, is supported by a NARSAD young investigator award, a CIHR (SVB397205) and Natural Science and Engineering Research Council (NSERC; RGPIN-2019-06496) grants and receives FRQS Junior-1 salary support; this work was also made possible by resources supported by the Quebec Network on Suicide, Mood Disorders and Related Disorders.
Author information
Authors and Affiliations
Contributions
BL, GT, NM and MJM planned the experiments, wrote the manuscript and supervised the studies. BL conducted and supervised all experiments and analysed all data. KA, TB, GM, VY and JY participated in the experiments. SAG, SJR and EJN provided help and support for the design and interpretation of the animal experiments. LN, DC, CG, JPL, RET and GC contributed to the experiments. RLN provided viral constructs.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Labonté, B., Abdallah, K., Maussion, G. et al. Regulation of impulsive and aggressive behaviours by a novel lncRNA. Mol Psychiatry 26, 3751–3764 (2021). https://doi.org/10.1038/s41380-019-0637-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-019-0637-4
This article is cited by
-
Techniques for investigating lncRNA transcript functions in neurodevelopment
Molecular Psychiatry (2023)
-
From warrior genes to translational solutions: novel insights into monoamine oxidases (MAOs) and aggression
Translational Psychiatry (2021)