Abstract
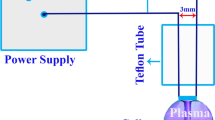
Plasma technology as an eco-friendly efficient strategy has gained much attention in various industries, especially in food, medicine, and agriculture. This study aimed to explore the cold plasma-mediated changes in growth, anatomy, expression of a WRKY1 transcription factor, and transcription rates of four key genes involved in the biosynthesis of cannabinoids (pharmaceutically valuable secondary metabolites) in hemp (Cannabis sativa L.). The seeds were treated with cold plasma (dielectric barrier discharge; 0.84 W cm−2; exposure times of 0, 40, and 80 s). The plasma treatment of 40 s increased biomass in both shoot and roots by an average of 57%, whereas the treatment at 80 s delayed growth and reduced it by 48%. Seed priming with plasma up-regulated the WRKY1 transcription factor (mean = 11.55 folds). Besides, the plasma treatments induced the expression of olivetolic acid cyclase by 42 folds. Furthermore, the plasma-primed seedlings also exhibited higher expression rates of olivetol synthase by 19 folds. With a similar trend, exposure to plasma stimulated transcription of cannabidiolic acid synthase by 12.4 folds. Up-regulations in Δ9-tetrahydrocannabinolic acid synthase also occurred following seed priming with plasma by 25.6 folds. Seed priming with plasma exhibits high potency to up-regulate expressions of genes involved in the productions of secondary metabolites, like cannabinoids. These results imply that the plasma reception and signal transduction can alter expressions of genes at the transcriptional level through which plasma priming may improve plant protection and secondary metabolism.






Similar content being viewed by others
References
Van Den Broeck HC, Maliepaard C, Ebskamp MJ, Toonen MA, Koops AJ (2008) Differential expression of genes involved in C1 metabolism and lignin biosynthesis in wooden core and bast tissues of fibre hemp (Cannabis sativa L.). Plant Sci 174(2):205–220. https://doi.org/10.1016/j.plantsci.2007.11.008
Flores-Sanchez IJ, Verpoorte R (2008) Secondary metabolism in cannabis. Phytochem Rev 7(3):615–639. https://doi.org/10.1007/s11101-008-9094-4
Gagne SJ, Stout JM, Liu E, Boubakir Z, Clark SM, Page JE (2012) Identification of olivetolic acid cyclase from Cannabis sativa reveals a unique catalytic route to plant polyketides. Proc Natl Acad Sci 109(31):12811–12816. https://doi.org/10.1073/pnas.1200330109
Taura F, Tanaka S, Taguchi C, Fukamizu T, Tanaka H, Shoyama Y, Morimoto S (2009) Characterization of olivetol synthase, a polyketide synthase putatively involved in cannabinoid biosynthetic pathway. FEBS Lett 583(12):2061–2066. https://doi.org/10.1016/j.febslet.2009.05.024
Taura F, Sirikantaramas S, Shoyama Y, Yoshikai K, Shoyama Y, Morimoto S (2007) Cannabidiolic-acid synthase, the chemotype-determining enzyme in the fiber-type Cannabis sativa. FEBS Lett 581(16):2929–2934. https://doi.org/10.1016/j.febslet.2007.05.043
Sirikantaramas S, Taura F, Morimoto S, Shoyama Y (2007) Recent advances in Cannabis sativa research: biosynthetic studies and its potential in biotechnology. Curr Pharm Biotechnol 8(4):237–243. https://doi.org/10.2174/138920107781387456
Happyana N, Kayser O (2016) Monitoring metabolite profiles of Cannabis sativa L. trichomes during flowering period using 1H NMR-based metabolomics and real-time PCR. Planta Med 82(13):1217–1223. https://doi.org/10.1055/s-0042-108058
Jalali S, Salami SA, Sharifi M, Sohrabi S (2019) Signaling compounds elicit expression of key genes in cannabinoid pathway and related metabolites in cannabis. Ind Crops Prod 133:105–110. https://doi.org/10.1016/j.indcrop.2019.03.004
Sheteiwy MS, An J, Yin M, Jia X, Guan Y, He F, Hu J (2019) Cold plasma treatment and exogenous salicylic acid priming enhances salinity tolerance of Oryza sativa seedlings. Protoplasma 256(1):79–99. https://doi.org/10.1007/s00709-018-1279-0
Seddighinia FS, Iranbakhsh A, Ardebili ZO, Satari TN, Soleimanpour S (2019) Seed priming with cold plasma and multi-walled carbon nanotubes modified growth, tissue differentiation, anatomy, and yield in bitter melon (Momordica charantia). J Plant Growth Regul. https://doi.org/10.1007/s00344-019-09965-2
Iranbakhsh A, Ardebili NO, Ardebili ZO, Shafaati M, Ghoranneviss M (2018) Non-thermal plasma induced expression of heat shock factor A4A and improved wheat (Triticum aestivum L.) growth and resistance against salt stress. Plasma Chem Plasma Process 38(1):29–44. https://doi.org/10.1007/s11090-017-9861-3
Iranbakhsh A, Ardebili ZO, Ardebili NO, Ghoranneviss M, Safari N (2018) Cold plasma relieved toxicity signs of nano zinc oxide in Capsicum annuum cayenne via modifying growth, differentiation, and physiology. Acta Physiol Plant 40(8):154. https://doi.org/10.1007/s11738-018-2730-8
Moghanloo M, Iranbakhsh A, Ebadi M, Ardebili ZO (2019) Differential physiology and expression of phenylalanine ammonia lyase (PAL) and universal stress protein (USP) in the endangered species Astragalus fridae following seed priming with cold plasma and manipulation of culture medium with silica nanoparticles. 3 Biotech 9(7):288. https://doi.org/10.1007/s13205-019-1822-5
Moghanloo M, Iranbakhsh A, Ebadi M, Satari TN, Ardebili ZO (2019) Seed priming with cold plasma and supplementation of culture medium with silicon nanoparticle modified growth, physiology, and anatomy in Astragalus fridae as an endangered species. Acta Physiol Plant 41(4):54. https://doi.org/10.1007/s11738-019-2846-5
Iranbakhsh A, Ghoranneviss M, Ardebili ZO, Ardebili NO, Tackallou SH, Nikmaram H (2017) Non-thermal plasma modified growth and physiology in Triticum aestivum via generated signaling molecules and UV radiation. Biol Plant 61(4):702–708. https://doi.org/10.1007/s10535-016-0699-y
Safari N, Iranbakhsh A, Ardebili ZO (2017) Non-thermal plasma modified growth and differentiation process of Capsicum annuum PP805 Godiva in in vitro conditions. Plasma Sci Technol 19(5):055501. https://doi.org/10.1088/2058-6272/aa57ef
Babajani A, Iranbakhsh A, Ardebili ZO, Eslami B (2019) Seed priming with non-thermal plasma modified plant reactions to selenium or zinc oxide nanoparticles: cold plasma as a novel emerging tool for plant science. Plasma Chem Plasma Process 39(1):21–34. https://doi.org/10.1007/s11090-018-9934-y
Mildažienė V, Aleknavičiūtė V, Žūkienė R, Paužaitė G, Naučienė Z, Filatova I, Lyushkevich V, Haimi P, Tamošiūnė I, Baniulis D (2019) Treatment of common sunflower (Helianthus annus L.) seeds with radio-frequency electromagnetic field and cold plasma induces changes in seed phytohormone balance, seedling development and leaf protein expression. Sci Rep 9(1):6437. https://doi.org/10.1038/s41598-019-42893-5
Ling L, Jiangang L, Minchong S, Chunlei Z, Yuanhua D (2015) Cold plasma treatment enhances oilseed rape seed germination under drought stress. Sci Rep 5:13033
Kyzek S, Holubová Ľ, Medvecká V, Tomeková J, Gálová E, Zahoranová A (2019) Cold atmospheric pressure plasma can induce adaptive response in pea seeds. Plasma Chem Plasma Process 39(2):475–486
Liu L, White MJ, MacRae TH (1999) Transcription factors and their genes in higher plants: functional domains, evolution and regulation. Eur J Biochem 262(2):247–257. https://doi.org/10.1046/j.1432-1327.1999.00349.x
Yu F, Huaxia Y, Lu W, Wu C, Cao X, Guo X (2012) GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L), is involved in disease resistance and plant development. BMC Plant Biol 12(1):144. https://doi.org/10.1186/1471-2229-12-144
Wang Q, Wang M, Zhang X, Hao B, Kaushik SK, Pan Y (2011) WRKY gene family evolution in Arabidopsis thaliana. Genetica 139(8):973. https://doi.org/10.1007/s10709-011-9599-4
Bakshi M, Oelmüller R (2014) WRKY transcription factors: jack of many trades in plants. Plant Signal Behav 9(2):e27700. https://doi.org/10.4161/psb.27700
Babajani A, Iranbakhsh A, Ardebili ZO, Eslami B (2019) Differential growth, nutrition, physiology, and gene expression in Melissa officinalis mediated by zinc oxide and elemental selenium nanoparticles. Environ Sci Pollut Res 26(24):24430–24444
Sera B, Sery M, Gavril B, Gajdova I (2017) Seed germination and early growth responses to seed pre-treatment by non-thermal plasma in hemp cultivars (Cannabis sativa L.). Plasma Chem Plasma Process 37(1):207–221. https://doi.org/10.1007/s11090-016-9763-9
Ji SH, Choi KH, Pengkit A, Im JS, Kim JS, Kim YH, Park Y, Hong EJ, Kyung Jung S, Choi EH, Park G (2016) Effects of high voltage nanosecond pulsed plasma and micro DBD plasma on seed germination, growth development and physiological activities in spinach. Arch Biochem Biophys 605:117–128
Shapira Y, Bormashenko E, Drori E (2019) Pre-germination plasma treatment of seeds does not alter cotyledon DNA structure, nor phenotype and phenology of tomato and pepper plants. Biochem Biophys Res Commun 519(3):512–517
Li C, He X, Luo X, Xu L, Liu L, Min L, Jin L, Zhu L, Zhang X (2014) Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol 166(4):2179–2194. https://doi.org/10.1104/pp.114.246694
Shinde BA, Dholakia BB, Hussain K, Aharoni A, Giri AP, Kamble AC (2018) WRKY1 acts as a key component improving resistance against Alternaria solani in wild tomato, Solanum arcanum Peralta. Plant Biotechnol J 16(8):1502–1513. https://doi.org/10.1111/pbi.12892
Heerah S, Katari M, Penjor R, Coruzzi G, Marshall-Colon A (2019) WRKY1 mediates transcriptional crosstalk between light and nitrogen signaling pathways in Arabidopsis thaliana. bioRxiv 1:603142
Safari M, Ardebili ZO, Iranbakhsh A (2018) Selenium nano-particle induced alterations in expression patterns of heat shock factor A4A (HSFA4A), and high molecular weight glutenin subunit 1Bx (Glu-1Bx) and enhanced nitrate reductase activity in wheat (Triticum aestivum L.). Acta Physiol Plant 40(6):117
Ren X, Chen Z, Liu Y, Zhang H, Zhang M, Liu Q, Hong X, Zhu JK, Gong Z (2010) ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J 63(3):417–429. https://doi.org/10.1111/j.1365-313X.2010.04248.x
Schluttenhofer C, Pattanaik S, Patra B, Yuan L (2014) Analyses of Catharanthus roseus and Arabidopsis thaliana WRKY transcription factors reveal involvement in jasmonate signaling. BMC Genom 15(1):502. https://doi.org/10.1186/1471-2164-15-502
Palmieri MC, Sell S, Huang X, Scherf M, Werner T, Durner J, Lindermayr C (2008) Nitric oxide-responsive genes and promoters in Arabidopsis thaliana: a bioinformatics approach. J Exp Bot 59(2):177–186. https://doi.org/10.1093/jxb/erm345
Kabir AH, Rahman MM, Das U, Sarkar U, Roy NC, Reza MA, Talukder MR, Uddin MA (2019) Reduction of cadmium toxicity in wheat through plasma technology. PLoS ONE 14(4):e0214509
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Iranbakhsh, A., Oraghi Ardebili, Z., Molaei, H. et al. Cold Plasma Up-Regulated Expressions of WRKY1 Transcription Factor and Genes Involved in Biosynthesis of Cannabinoids in Hemp (Cannabis sativa L.). Plasma Chem Plasma Process 40, 527–537 (2020). https://doi.org/10.1007/s11090-020-10058-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11090-020-10058-2