Abstract
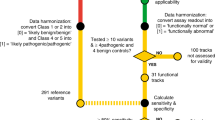
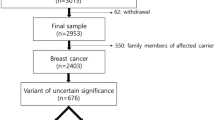
The clinical utility of BRCA1/2 genotyping was recently extended from the selection of subjects at high risk for hereditary breast and ovary cancer to the identification of candidates for poly (ADP-ribose) polymerase (PARP) inhibitor treatment. This underscores the importance of accurate interpretation of BRCA1/2 genetic variants and of reducing the number of variants of uncertain significance (VUSs). Two recent studies by Findlay et al. and Starita et al. introduced high-throughput functional assays, and proactively analyzed variants in specific regions regardless of whether they had been previously observed. We retrospectively reviewed all BRCA1 and BRCA2 germline genetic test reports from patients with breast or ovarian cancer examined at Asan Medical Center (Seoul, Korea) between September 2011 and December 2018. Variants were assigned pathogenic or benign strong evidence codes according to the functional classification and were reclassified according to the ACMG/AMP 2015 guidelines. Among 3684 patients with available BRCA1 and BRCA2 germline genetic test reports, 429 unique variants (181 from BRCA1) were identified. Of 34 BRCA1 variants intersecting with the data reported by Findlay et al., three missense single-nucleotide variants from four patients (0.11%, 4/3684) were reclassified from VUSs to likely pathogenic variants. Four variants scored as functional were reclassified into benign or likely benign variants. Three variants that overlapped with the data reported by Starita et al. could not be reclassified. In conclusion, proactive high-throughput functional study data are useful for the reclassification of clinically observed VUSs. Integrating additional evidence, including functional assay results, may help reduce the number of VUSs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Daly MB, Pilarski R, Berry M, Buys SS, Farmer M, Friedman S, et al. NCCN guidelines insights: genetic/familial high-risk assessment: breast and ovarian, version 2.2017. J Natl Compr Canc Netw. 2017;15:9–20.
Pujade-Lauraine E, Ledermann JA, Selle F, Gebski V, Penson RT, Oza AM, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol 2017;18:1274–84.
Golubeva VA, Nepomuceno TC, Monteiro ANA. Germline missense variants in BRCA1: new trends and challenges for clinical annotation. Cancers. 2019;11:522.
Toland AE, Andreassen PR. DNA repair-related functional assays for the classification of BRCA1 and BRCA2 variants: a critical review and needs assessment. J Med Genet. 2017;54:721–31.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Morris JR, Pangon L, Boutell C, Katagiri T, Keep NH, Solomon E. Genetic analysis of BRCA1 ubiquitin ligase activity and its relationship to breast cancer susceptibility. Hum Mol Genet. 2006;15:599–606.
Phelan CM, Dapic V, Tice B, Favis R, Kwan E, Barany F, et al. Classification of BRCA1 missense variants of unknown clinical significance. J Med Genet. 2005;42:138–46.
Lee MS, Green R, Marsillac SM, Coquelle N, Williams RS, Yeung T, et al. Comprehensive analysis of missense variations in the BRCT domain of BRCA1 by structural and functional assays. Cancer Res. 2010;70:4880–90.
Bouwman P, van der Gulden H, van der Heijden I, Drost R, Klijn CN, Prasetyanti P, et al. A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov. 2013;3:1142–55.
Ransburgh DJ, Chiba N, Ishioka C, Toland AE, Parvin JD. Identification of breast tumor mutations in BRCA1 that abolish its function in homologous DNA recombination. Cancer Res. 2010;70:988–95.
Kuznetsov SG, Liu P, Sharan SK. Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nat Med. 2008;14:875–81.
Parsons MT, Tudini E, Li H, Hahnen E, Wappenschmidt B, Feliubadalo L, et al. Large scale multifactorial likelihood quantitative analysis of BRCA1 and BRCA2 variants: an ENIGMA resource to support clinical variant classification. Hum Mutat. 2019;40:1557–78.
Starita LM, Islam MM, Banerjee T, Adamovich AI, Gullingsrud J, Fields S, et al. A multiplex homology-directed DNA repair assay reveals the impact of more than 1,000 BRCA1 missense substitution variants on protein function. Am J Hum Genet. 2018;103:498–508.
Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217–22.
Shin Y, Choi C, Lee J, Shin SY. First step to big data research in hospital. Stud Health Technol Inform. 2015;216:924.
Jhuraney A, Velkova A, Johnson RC, Kessing B, Carvalho RS, Whiley P, et al. BRCA1 Circos: a visualisation resource for functional analysis of missense variants. J Med Genet. 2015;52:224–30.
Cline MS, Liao RG, Parsons MT, Paten B, Alquaddoomi F, Antoniou A, et al. BRCA Challenge: BRCA Exchange as a global resource for variants in BRCA1 and BRCA2. PLoS Genet. 2018;14:e1007752.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019. https://doi.org/10.1101/531210:531210.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81.
Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–2.
Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67.
Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–94.
Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–21.
Easton DF, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, Allen-Brady K, et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007;81:873–83.
Lee JS, Oh S, Park SK, Lee MH, Lee JW, Kim SW, et al. Reclassification of BRCA1 and BRCA2 variants of uncertain significance: a multifactorial analysis of multicentre prospective cohort. J Med Genet. 2018;55:794–802.
Biesecker LG, Harrison SM. The ACMG/AMP reputable source criteria for the interpretation of sequence variants. Genet Med. 2018;20:1687–8.
Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:1062–7.
Vallee MP, Di Sera TL, Nix DA, Paquette AM, Parsons MT, Bell R, et al. Adding in silico assessment of potential splice aberration to the integrated evaluation of BRCA gene unclassified variants. Hum Mutat. 2016;37:627–39.
Plon SE, Eccles DM, Easton D, Foulkes WD, Genuardi M, Greenblatt MS, et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29:1282–91.
McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–70.
Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43:295–305.
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:886–94.
Kang E, Park SK, Lee JW, Kim Z, Noh WC, Jung Y, et al. KOHBRA BRCA risk calculator (KOHCal): a model for predicting BRCA1 and BRCA2 mutations in Korean breast cancer patients. J Hum Genet. 2016;61:365–71.
Frank TS, Deffenbaugh AM, Reid JE, Hulick M, Ward BE, Lingenfelter B, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–90.
Slavin TP, Manjarrez S, Pritchard CC, Gray S, Weitzel JN. The effects of genomic germline variant reclassification on clinical cancer care. Oncotarget. 2019;10:417–23.
Moghadasi S, Eccles DM, Devilee P, Vreeswijk MP, van Asperen CJ. Classification and clinical management of variants of uncertain significance in high penetrance cancer predisposition genes. Hum Mutat. 2016;37:331–6.
Eccles BK, Copson E, Maishman T, Abraham JE, Eccles DM. Understanding of BRCA VUS genetic results by breast cancer specialists. BMC Cancer. 2015;15:936.
Makhnoon S, Garrett LT, Burke W, Bowen DJ, Shirts BH. Experiences of patients seeking to participate in variant of uncertain significance reclassification research. J Community Genet. 2019;10:189–96.
Guidugli L, Shimelis H, Masica DL, Pankratz VS, Lipton GB, Singh N, et al. Assessment of the clinical relevance of BRCA2 missense variants by functional and computational approaches. Am J Hum Genet. 2018;102:233–48.
Guidugli L, Pankratz VS, Singh N, Thompson J, Erding CA, Engel C, et al. A classification model for BRCA2 DNA binding domain missense variants based on homology-directed repair activity. Cancer Res. 2013;73:265–75.
Brnich SE, Rivera-Munoz EA, Berg JS. Quantifying the potential of functional evidence to reclassify variants of uncertain significance in the categorical and Bayesian interpretation frameworks. Hum Mutat. 2018;39:1531–41.
Nykamp K, Anderson M, Powers M, Garcia J, Herrera B, Ho YY, et al. Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med. 2017;19:1105–17.
Kang E, Seong MW, Park SK, Lee JW, Lee J, Kim LS, et al. The prevalence and spectrum of BRCA1 and BRCA2 mutations in Korean population: recent update of the Korean Hereditary Breast Cancer (KOHBRA) study. Breast Cancer Res Treat. 2015;151:157–68.
Kwon BS, Byun JM, Lee HJ, Jeong DH, Lee TH, Shin KH, et al. Clinical and genetic characteristics of BRCA1/2 mutation in Korean ovarian cancer patients: a multicenter study and literature review. Cancer Res Treat. 2019;51:941–50.
Schwartz GF, Hughes KS, Lynch HT, Fabian CJ, Fentiman IS, Robson ME, et al. Proceedings of the international consensus conference on breast cancer risk, genetics, & risk management, April, 2007. Breast J. 2009;15:4–16.
Rebbeck TR, Friebel TM, Friedman E, Hamann U, Huo D, Kwong A, et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum Mutat. 2018;39:593–620.
Hoskinson DC, Dubuc AM, Mason-Suares H. The current state of clinical interpretation of sequence variants. Curr Opin Genet Dev. 2017;42:33–9.
Fernandes VC, Golubeva VA, Di Pietro G, Shields C, Amankwah K, Nepomuceno TC, et al. Impact of amino acid substitutions at secondary structures in the BRCT domains of the tumor suppressor BRCA1: Implications for clinical annotation. J Biol Chem. 2019;294:5980–92.
Brnich SE, Abou Tayoun AN, Couch FJ, Cutting G, Greenblatt MS, Heinen CD, et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. bioRxiv. 2019. https://doi.org/10.1101/709428:709428.
Acknowledgements
We would like to thank Asan Medical Library for the paper formatting.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, HK., Lee, E.J., Lee, YJ. et al. Impact of proactive high-throughput functional assay data on BRCA1 variant interpretation in 3684 patients with breast or ovarian cancer. J Hum Genet 65, 209–220 (2020). https://doi.org/10.1038/s10038-019-0713-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0713-2