Abstract
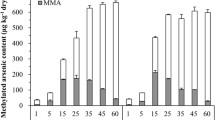
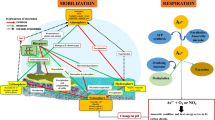
Microbial arsenic (As) methylation plays important roles in the As biogeochemical cycle. However, little is known about the diversity and functions of As-methylating microorganisms from the tailings of a Realgar Mine, which is characterized as containing extremely high concentrations of As. To address this issue, we collected five samples (T1–T5) from the tailings of Shimen Realgar Mine. Microcosm assays without addition of exogenous As and carbon indicated that all the five samples possess significant As-methylating activities, producing 0.8–5.7 μg/L DMAsV, and 1.1–10.7 μg/L MMAsV with an exception of T3, from which MMAsV was not detectable after 14.0 days of incubation. In comparison, addition of 20.0 mM lactate to the microcosms significantly enhanced the activities of these samples; the produced DMAsV and MMAsV are 8.0–39.7 μg/L and 5.8–38.3 μg/L, respectively. The biogenic DMAsV shows significant positive correlations with the Fe concentrations and negative correlations with the total nitrogen concentrations in the environment. A total of 63 different arsM genes were identified from the five samples, which code for new or new-type ArsM proteins, suggesting that a unique diversity of As-methylating microbes are present in the environment. The microbial community structures of the samples were significantly shaped by the environmental total organic carbon, total As contents and NO3− contents. These data help to better understand the microorganisms-catalyzed As methylation occurred in the environment with extremely high contents of As.






Similar content being viewed by others
References
Ajees AA, Marapakala K, Packianathan C, Sankaran B, Rosen BP (2012) Structure of an As(III) S-adenosylmethionine methyltransferase: insights into the mechanism of arsenic biotransformation. Biochemistry 51:5476–5485
Bagade AV, Bachate SP, Dholakia BB, Giri AP, Kodam KM (2016) Characterization of Roseomonas and Nocardioides spp. for arsenic transformation. J Hazard Mater 318:742–750
Bhattacharya P, Welch AH, Stollenwerk KG, McLaughlin MJ, Bundschuh J, Panaullah G (2007) Arsenic in the environment: biology and chemistry. Sci Total Environ 379(2):109–120
Broderick KE, Singh V, Zhuang S, Kambo A, Chen JC, Sharma VS, Pilz RB, Boss GR (2005) Nitric oxide scavenging by the coba-613 lamin precursor cobinamide. J Biol Chem 280:8678–8685
Cai L, Yu K, Yang Y, Chen BW, Li XD, Zhang T (2013) Metagenomic exploration reveals high levels of microbial arsenic metabolism genes in activated sludge and coastal sediments. Appl Microbiol Biotechnol 97:9579–9588
Chen J, Sun GX, Wang XX, Lorenzo VD, Rosen BP, Zhu YG (2014) Volatilization of arsenic from polluted soil by Pseudomonas putida engineered for expression of the arsM arsenic(III) S-adenosine methyltransferase gene. Environ Sci Technol 48:10337–10344
Chen X, Zeng XC, Wang J, Deng Y, Ma T, Guoji E et al. (2017) Microbial communities involved in arsenic mobilization and release from the ndeep sediments into groundwater in Jianghan plain, Central China. Sci Total Environ 579:989–999
Fisher JC, Hollibaugh JT (2008) Selenate-dependent anaerobic arsenite oxidation by a bacterium from Mono Lake, California. Appl Environ Microbiol 74:2588–2594
Huang K, Chen C, Zhang J, Tang Z, Shen Q, Rosen BP et al. (2016) Efficient arsenic methylation and volatilization mediated by a novel bacterium from an arsenic-contaminated paddy soil. Environ Sci Technol 50:6389–6396
Huang K, Xu Y, Zhang J, Chen C, Gao F, Zhao FJ (2017) Arsenicibacter rosenii gen. nov. sp. nov. an efficient arsenic methylating and volatilizing bacterium isolated from an arsenic-contaminated paddy soil. Int J Syst Evol Microbiol 67:3186–3191
Jia Y, Huang H, Zhong M, Wang FH, Zhang LM, Zhu YG (2013) Microbial arsenic methylation in soil and rice rhizosphere. Environ Sci Technol 47:3141–3148
Kim DJ, Lee DI, Keller J (2006) Effect of temperature and free ammonia on nitrification and nitrite accumulation in landfill leachate and analysis of its nitrifying bacterial community by FISH. Bioresour Technol 97:459–468
Kudo K, Yamaguchi N, Makino T, Ohtsuka T, Kimura K, Dong DT et al. (2013) Release of arsenic from soil by a novel dissimilatory arsenate-reducing bacterium, Anaeromyxobacter sp. strain PSR-1. Appl Environ Microbiol 79:4635–4642
Kulp TR (2014) Early earth: arsenic and primordial life. Nat Geosci 7:785–786
Kuramata M, Sakakibara F, Kataoka R, Abe T, Asano M, Baba K et al. (2015) Arsenic biotransformation by Streptomyces sp. isolated from rice rhizosphere. Environ Microbiol 17:1897–1909
Lear G, Song B, Gault AG, Polya DA, Lloyd JR (2007) Molecular analysis of arsenate-reducing bacteria within Cambodian sediments following amendment with acetate. Appl Environ Microbiol 73:1041–1048
Li H, Zeng XC, He Z, Chen X, Guoji E, Han Y, Wang Y (2016) Long-term performance of rapid oxidation of arsenite in simulated groundwater using a population of arsenite-oxidizing microorganisms in a bioreactor. Water Res 101:393–401
Maguffin SC, Kirk MF, Daigle AR, Hinkle SR, Jin Q (2015) Substantial contribution of biomethylation to aquifer arsenic cycling. Nat Geosci 8(4):290–293
Mestrot A, Planer-Friedrich B, Feldmann J (2013) Biovolatilisation: a poorly studied pathway of the arsenic biogeochemical cycle. Environ. Sci. Process Impacts 15:1639–1651
Nordstrom DK (2002) Public health: worldwide occurrences of arsenic in ground water. Science 296:2143–2145
Ohtsuka T, Yamaguchi N, Makino T, Sakurai K, Kimura K, Kudo K et al. (2013) Arsenic dissolution from Japanese paddy soil by a dissimilatory arsenate-reducing bacterium Geobacter sp. OR-1. Environ Sci Technol 47:6263–6271
Oremland RS, Stolz JF (2003) The ecology of arsenic. Science 300:939–944
Osborne Kulp TH, McArthur JM, Sikdar PK, Santini JM (2015) Isolation of an arsenate-respiring bacterium from a redox front in an arsenic-polluted aquifer in West Bengal, Bengal Basin. Environ Sci Technol 49:4193–4199
Qin J, Lehr CR, Yuan C, Le XC, McDermott TR, Rosen BP (2009) Biotransformation of arsenic by a Yellowstone thermoacidophilic eukaryotic alga. Proc Natl Acad Sci USA 106:5213–5217
Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C (2006) Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci USA 103:2075–2080
Reid MC, Maillard J, Bagnoud A, Falquet L, Le VP, Bernierlatmani R (2017) Arsenic methylation dynamics in a rice paddy soil anaerobic enrichment culture. Environ Sci Technol 51:10546–10554
Rhine ED, Onesios KM, Serfes ME, Reinfelder JR, Young LY (2008) Arsenic transformation and mobilization from minerals by the arsenite oxidizing strain WAO. Environ Sci Technol 42:1423–1429
Schaefer MV, Ying SC, Benner SG, Duan Y, Wang Y, Fendorf S (2016) Aquifer arsenic cycling induced by seasonal hydrologic changes within the Yangtze River basin. Environ Sci Technol 50:3521–3529
Shi W, Wu W, Zeng XC, Chen X, Zhu X, Cheng S (2018) Dissimilatory arsenate-respiring prokaryotes catalyze the dissolution, reduction and release of arsenic from paddy soils into groundwater: implication for the effect of sulfate. Ecotoxicology 27:1126–1136
Singh R, Singh S, Parihar P, Singh V, Prasad S (2015) Arsenic contamination, consequences and remediation techniques: a review. Ecotoxicol Environ Saf 112:247–270
Slyemi D, Bonnefoy V (2012) How prokaryotes deal with arsenic. Environ Microbiol Rep. 4:571–586
Song B, Chyun E, Jaffé PR, Ward BB (2009) Molecular methods to detect and monitor dissimilatory arsenate-respiring bacteria (DARB) in sediments. FEMS Microb Ecol 68:108–117
Srivastava PK, Vaish A, Dwivedi S, Chakrabarty D, Singh N, Tripathi RD (2011) Biological removal of arsenic pollution by soil fungi. Sci Total Environ 409:2430–2442
Sun W, Sierraalvarez R, Field JA (2011) Long term performance of an arsenite-oxidizing-chlorate-reducing microbial consortium in an upflow anaerobic sludge bed (uasb) bioreactor. Bioresour Technol 102:5010–5016
Wang J, Wu M, Gan L, Si Y (2016) Biotransformation and biomethylation of arsenic by Shewanella oneidensis MR-1. Chemosphere 145:329–335
Wang J, Zeng XC, Zhu X, Chen X, Zeng X, Mu Y (2017) Sulfate enhances the dissimilatory arsenate-respiring prokaryotes-mediated mobilization, reduction and release of insoluble arsenic and iron from the arsenic-rich sediments into groundwater. J Hazard Mater 339:409–417
Wang PP, Bao P, Sun GX (2015) Identification and catalytic residues of the arsenite methyltransferase from a sulfate-reducing bacterium, Clostridium sp. BXM. FEMS Microbiol Lett 362:1–8
Wang PP, Sun GX, Zhu YG (2014) Identification and characterization of the arsenite methyltransferase from an archaeon, Methanosarcina acetivorans C2A. Environ Sci Technol 48(21):12706–12713
Wu X, Zhang H, Chen J et al. (2016) Comparison of the fecal microbiota of dholes high-throughput Illumina sequencing of the V3–V4 region of the 16S rRNA gene. Appl Microbiol Biotechnol 100(8):3577–3586
Xue XM, Ye J, Raber G et al. (2017) Arsenic methyltransferase is involved in arsenosugar biosynthesis by providing dma. Environ. Sci. Technol 51:1224–1230
Yang Y, Mu Y, Zeng XC, Wu W, Yuan J, Liu Y et al. (2017) Functional genes and thermophilic microorganisms responsible for arsenite oxidation from the shallow sediment of an untraversed hot spring outlet. Ecotoxicology 26:490–501
Ye J, Chang Y, Yan Y et al. (2014) Identification and characterization of the arsenite methyltransferase from a protozoan, Tetrahymena pyriformis. Aquat Toxicol 149(1):50–57
Zeng XC, EG, Wang J, Wang N, Chen X, Mu Y, Li H, Yang Y, Liu Y, Wang Y (2016) Functions and unique diversity of genes and microorganisms involved in arsenite oxidation from the tailings of a realgar mine Appl Environ Microbiol 82:7019–7029
Zeng XC, He Z, Chen X, Cao QAD, Li H, Wang Y (2018a) Effects of arsenic on the biofilm formations of arsenite-oxidizing bacteria. Ecotoxicol Environ Saf 165:1–10
Zeng XC, Yang Y, Shi W, Peng Z, Chen X, Zhu X, Wang Y (2018b) Microbially mediated methylation of arsenic in the arsenic-rich soils and sediments of Jianghan plain. Front Microbiol 9:1389
Zhai W, Wong MT, Luo F, Hashmi MZ, Liu X, Edwards EA et al. (2017) Arsenic methylation and its relationship to abundance and diversity of ARSM genes in composting manure. Sci Rep. 7:42198
Zhang J, Cao T, Tang Z, Shen Q, Rosen BP, Zhao FJ (2015b) Arsenic methylation and volatilization by arsenite S-adenosylmethionine methyltransferase in Pseudomonas alcaligenes NBRC14159. Appl Environ Microbiol 81:2852–2860
Zhang J, Zhou W, Liu B, He J, Shen Q, Zhao FJ (2015a) Anaerobic arsenite oxidation by an autotrophic arsenite-oxidizing bacterium from an arsenic-contaminated paddy soil. Environ Sci Technol 49:5956–5964
Zhang SY, Su JQ, Sun GX, Yang Y, Zhao Y, Ding J et al. (2017) Land scale biogeography of arsenic biotransformation genes in estuarine wetland. Environ Microbiol 19:2468–2482
Zhu X, Wang R, Lu X, Liu H, Li J, Ouyang B, Lu J (2015) Secondary minerals of weathered orpiment-realgar-bearing tailings in Shimen carbonate-type realgar mine, Changde, Central China. Miner Petrol 109:1–15
Zhu X, Zeng XC, Chen X, Wu W, Wang Y (2019) Inhibitory effect of nitrate/nitrite on the microbial reductive dissolution of arsenic and iron from soils into pore water. Ecotoxicology 28:528–538
Zhu YG, Xue XM, Kappler A, Rosen BP, Meharg AA (2017) Linking genes to microbial biogeochemical cycling: lessons from arsenic. Environ Sci Technol 51:7326–7339
Zhu YG, Yoshinaga M, Zhao FJ, Rosen BP (2014) Earth abides arsenic biotransformations. Annu Rev Earth Planet Sci 42:443–467
Acknowledgements
This work was supported by the General Programs and the Foundations for Innovative Research Groups from the National Natural Science Foundation of China (grant nos. 41472257, 41521001), and the Fundamental Research Funds for the Central Universities, China University of Geosciences (Wuhan) (grant no. CUGCJ1702).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the author.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ngegla, J.V., Zhou, X., Chen, X. et al. Unique diversity and functions of the arsenic-methylating microorganisms from the tailings of Shimen Realgar Mine. Ecotoxicology 29, 86–96 (2020). https://doi.org/10.1007/s10646-019-02144-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10646-019-02144-9