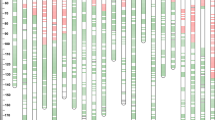
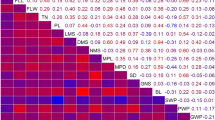
Abstract
Key message
Eight environmentally stable QTL for grain yield-related traits were detected by four RIL populations, and two of them were validated by a natural wheat population containing 580 diverse varieties or lines.
Abstract
Yield and yield-related traits are important factors in wheat breeding. In this study, four RIL populations derived from the cross of one common parent Yanzhan 1 (a Chinese domesticated cultivar) and four donor parents including Hussar (a British domesticated cultivar) and three semi-wild wheat varieties in China were phenotyped for 11 yield-related traits in eight environments. An integrated genetic map containing 2009 single-nucleotide polymorphism (SNP) markers generated from a 90 K SNP array was constructed to conduct quantitative trait loci (QTL) analysis. A total of 161 QTL were identified, including ten QTL for grain yield per plant (GYP) and yield components, 49 QTL for spike-related traits, 43 QTL for flag leaf-related traits, 22 QTL for plant height (PH), and 37 QTL for heading date and flowering date. Eight environmentally stable QTL were validated in individual RIL population where the target QTL was notably detected, and six of them had a significant effect on GYP. Furthermore, Two QTL, QSPS-2A.4 and QSL-4A.1, were also validated in a natural wheat population containing 580 diverse varieties or lines, which provided valuable resources for further fine mapping and genetic improvement in yield in wheat.




Similar content being viewed by others
Abbreviations
- RIL:
-
Recombinant inbred line
- QTL:
-
Quantitative trait locus/loci
- SNP:
-
Single-nucleotide polymorphism
- MAS:
-
Marker-assisted selection
- BLUP:
-
Best linear unbiased prediction
- JICIM:
-
Joint inclusive composite interval mapping
- ANOVA:
-
Analysis of variance
- PVE:
-
Phenotypic variation explained
- GY:
-
Grain yield
- GYP:
-
Grain yield per plant
- SN:
-
Spike number per plant
- KPS:
-
Kernel number per spike
- SL:
-
Spike length
- SPS:
-
Spikelet number per spike
- FLL:
-
Flag leaf length
- FLW:
-
Flag leaf width
- FLA:
-
Flag leaf area
- PH:
-
Plant height
- HD:
-
Heading date
- FD:
-
Flowering date
- YZ:
-
Yanzhan 1
- CY:
-
Chayazheda29
- HU:
-
Hussar
- YN:
-
Yunnanxiaomai
- YT:
-
Yutiandaomai
References
Assanga SO, Fuentealba M, Zhang GR et al (2017) Mapping of quantitative trait loci for grain yield and its components in a US popular winter wheat TAM 111 using 90 K SNPs. PLoS ONE 12:e0189669
Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) A Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115:721–733
Chen F, Yu YX, Xia XC, He ZH (2007) Prevalence of a novel puroindoline b allele in Yunnan endemic wheats (Triticum aestivum ssp. yunnanense King). Euphytica 156:39–46
Chen SL, Gao RH, Wang HY et al (2015) Characterization of a novel reduced height gene (Rht23) regulating panicle morphology and plant architecture in bread wheat. Euphytica 203:583–594
Cui F, Ding AM, Li J et al (2012) QTL detection of seven spike-related traits and their genetic correlations in wheat using two related RIL populations. Euphytica 186:177–192
Cui F, Zhao C, Ding A, Li J, Wang L, Li X, Bao Y, Li J, Wang H (2014) Construction of an integrative linkage map and QTL mapping of grain yield-related traits using three related wheat RIL populations. Theor Appl Genet 127:659
Deng SM, Wu XR, Wu YY et al (2011) Characterization and precise mapping of a QTL increasing spike number with pleiotropic effects in wheat. Theor Appl Genet 122:281–289
Deng ZY, Cui Y, Han QD et al (2017) Discovery of consistent QTLs of wheat spike-related traits under nitrogen treatment at different development stages. Front Plant Sci 8:2120
Distelfeld A, Li C, Dubcovsky J (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12:178–184
Edae EA, Byrne PF, Haley SD et al (2014) Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes. Theor Appl Genet 127:791–807
Fan XL, Cui F, Zhao CH et al (2015) QTLs for flag leaf size and their influence on yield-related traits in wheat (Triticum aestivum L.). Mol Breed 35:24
Ford BA, Foo E, Sharwood R et al (2018) Rht18 semi-dwarfism in wheat is due to increased GA 2-oxidaseA9 expression and reduced GA content. Plant Physiol 177:168–180
Griffiths S, Simmonds J, Leverington M, Wang YK, Fish L, Sayers L, Alibert L, Orford S, Wingen L, Herry L, Faure S, Laurie D, Bilham L, Snape J (2009) Meta-QTL analysis of the genetic control of ear emergence in elite European winter wheat germplasm. Theor Appl Genet 119:383–395
Hedden P (2003) The genes of the green revolution. Trends Genet 19:5–9
Huang XQ, Kempf H, Ganal MW, Röder MS (2004) Advanced backcross QTL analysis in progenies derived from a cross between a German elite winter wheat variety and a synthetic wheat (Triticum aestivum L.). Theor Appl Genet 109:933–943
Huang M, Mheni N, Brown-Guedira G, Mckendry A, Griffey C, Sanford DV, Costa J, Sneller C (2018) Genetic analysis of heading date in winter and spring wheat. Euphytica 214:128
Huehn M (2010) Random variability of map distances based on Kosambi’s and Haldane’s mapping functions. J Appl Genet 51:27–31
Hussain W, Baenziger PS, Belamkar V et al (2017) Genotyping-by-sequencing derived high-density linkage map and its application to QTL mapping of flag leaf traits in bread wheat. Sci Rep 7:16394
Jia HY, Wan HS, Yang SH et al (2013) Genetic dissection of yield-related traits in a recombinant inbred line population created using a key breeding parent in China’s wheat breeding. Theor Appl Genet 126:2123–2139
Johnson EB, Nalam VJ, Zemetra RS, Riera-Lizarazu O (2008) Mapping the compactum locus in wheat (Triticum aestivum L.) and its relationship to other spike morphology genes of the Triticeae. Euphytica 163:193–201
Jr FEH (2015) Hmisc: Harrell Miscellaneous. R package version 3.16-0
Kosambi DD (2012) The estimation of map distances from recombination values. Ann Hum Genet 12:172–175
Ladizinsky G (1985) Founder effect in crop-plant evolution. Econ Bot 39:191–199
Li WL, Nelson JC, Chu CY et al (2002) Chromosomal locations and genetic relationship of tiller and spike characters in wheat. Euphytica 125:357–366
Li SS, Jia JZ, Wei XY et al (2007) A intervarietal genetic map and QTL analysis for yield traits in wheat. Mol Breed 20:167–178
Li FJ, Wen WE, He ZH et al (2018) Genome-wide linkage mapping of yield-related traits in three Chinese bread wheat populations using high-density SNP markers. Theor Appl Genet 131:1903–1924
Lian JF, Zhang DQ, Wu BJ et al (2016) QTL mapping of flag leaf traits using an integrated high-density 90 K genotyping chip. J Triticeae Crops 36:689–698
Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28:2397–2399
Liu G, Xu SB, Ni ZF et al (2011) Molecular dissection of plant height QTLs using recombinant inbred lines from hybrids between common wheat (Triticum aestivum L.) and spelt wheat (Triticum spelta L.). Chin Sci Bull 56:1897–1903
Liu K, Sun XX, Ning TY et al (2018a) Genetic dissection of wheat panicle traits using linkage analysis and a genome-wide association study. Theor Appl Genet 2:1–18
Liu YX, Tao Y, Wang ZQ et al (2018b) Identification of QTL for flag leaf length in common wheat and their pleiotropic effects. Mol Breed 38:11
Liu KY, Xu H, Liu G et al (2018c) QTL mapping of flag leaf-related traits in wheat (Triticum aestivum L.). Theor Appl Genet 131:839–849
Maccaferri M, Sanguineti MC, Demontis A et al (2011) Association mapping in durum wheat grown across a broad range of water regimes. J Exp Bot 62:409–438
Marza F, Bai GH, Carver BF, Zhou WC (2006) Quantitative trait loci for yield and related traits in the wheat population Ning7840 x Clark. Theor Appl Genet 112:688–698
Meng L, Li HH, Zhang LY, Wang JK (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3:269–283
Milner SG, Maccaferri M, Huang BE et al (2016) A multiparental cross population for mapping QTL for agronomic traits in durum wheat (Triticum turgidum ssp. durum). Plant Biotechnol J 14:735–748
Mo YJ, Vanzetti LS, Hale I et al (2018) Identification and characterization of Rht25, a locus on chromosome arm 6AS affecting wheat plant height, heading time, and spike development. Theor Appl Genet 131:2021–2035
Narasimhamoorthy B, Gill BS, Fritz AK et al (2006) Advanced backcross QTL analysis of a hard winter wheat x synthetic wheat population. Theor Appl Genet 112:787–796
Peng ZS, Li X, Yang ZJ, Liao ML (2011) A new reduced height gene found in the tetraploid semi-dwarf wheat landrace Aiganfanmai. Genet Mol Res 10:2349–2357
Quarrie SA, Pekic-Quarrie S, Radosevic R et al (2006) Dissecting a wheat QTL for yield present in a range of environments: from the QTL to candidate genes. J Exp Bot 57:2627–2637
Ren DQ, Fang XJ, Jiang P et al (2018) Genetic architecture of nitrogen-deficiency tolerance in wheat seedlings based on a nested association mapping (NAM) population. Front Plant Sci 9:845
Sakuma S, Golan G, Guo ZF et al (2019) Unleashing floret fertility in wheat through the mutation of a homeobox gene. PNAS 116:5182–5187
Schnaithmann F, Kopahnke D, Pillen K (2014) A first step toward the development of a barley NAM population and its utilization to detect QTLs conferring leaf rust seedling resistance. Theor Appl Genet 127:1513–1525
Simmonds J, Scott P, Leverington-Waite M et al (2014) Identification and independent validation of a stable yield and thousand grain weight QTL on chromosome 6A of hexaploid wheat (Triticum aestivum L.). BMC Plant Biol 14:191
Sourdille P, Tixier MH, Charmet G et al (2000) Location of genes involved in ear compactness in wheat (Triticum aestivum) by means of molecular markers. Mol Breed 6:247–255
Van-Ooijen JW (2006) JoinMapR4. Software for the calculation of genetic linkage maps in experimental populations, Wageningen, Holland
Vikhe P, Patil R, Chavan A et al (2017) Mapping gibberellin-sensitive dwarfing locus Rht18 in durum wheat and development of SSR and SNP markers for selection in breeding. Mol Breed 37:28
Wang HY, Wang XE, Chen PD, Liu DJ (2007) Progress on the study of Yunnan, Tibetan and Xinjiang wheat. J Triticeae Crops 27:740–743
Wang JS, Liu WH, Wang H et al (2011a) QTL mapping of yield-related traits in the wheat germplasm 3228. Euphytica 177:277–292
Wang P, Zhou GL, Yu HH, Yu SB (2011b) Fine mapping a major QTL for flag leaf size and yield-related traits in rice. Theor Appl Genet 123:1319–1330
Wang SC, Wong D, Forrest K et al (2014) Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Wang X, Dong L, Hu J, Pang Y, Hu L, Xiao G, Ma X, Kong X, Jia J, Wang H (2019) Dissecting genetic loci affecting grain morphological traits to improve grain weight via nested association mapping. Theor Appl Genet 132:3115–3128
Wilde F, Schön CC, Korzun V et al (2008) Marker-based introduction of three quantitative-trait loci conferring resistance to Fusarium head blight into an independent elite winter wheat breeding population. Theor Appl Genet 117:29–35
Wu QH, Chen YX, Fu L et al (2016) QTL mapping of flag leaf traits in common wheat using an integrated high-density SSR and SNP genetic linkage map. Euphytica 208:337–351
Xu YF, Wang RF, Tong YP et al (2014) Mapping QTLs for yield and nitrogen-related traits in wheat: influence of nitrogen and phosphorus fertilization on QTL expression. Theor Appl Genet 127:59–72
Xue SL, Xu F, Li GQ et al (2013) Fine mapping TaFLW1, a major QTL controlling flag leaf width in bread wheat (Triticum aestivum L.). Theor Appl Genet 126:1941–1949
Yang DL, Liu Y, Cheng HB et al (2016) Genetic dissection of flag leaf morphology in wheat (Triticum aestivum L.) under diverse water regimes. BMC Genet 17:94
Yuan QQ, Deng ZY, Peng T, Tian JC (2012) QTL-based analysis of heterosis for number of grains per spike in wheat using DH and immortalized F2 populations. Euphytica 188:387–395
Zeng XQ, Wang YJ, Li WY et al (2010) Comparison of the genetic diversity between Triticum aestivum ssp. tibetanum Shao and Tibetan wheat landraces (Triticum aestivum L.) by using intron-splice junction primers. Genet Resour Crop Evol 57:1141–1150
Zhai HJ, Feng ZY, Li J et al (2016) QTL analysis of spike morphological traits and plant height in winter wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Front Plant Sci 7:1617
Zhang JJ, Dell B, Biddulph B et al (2013) Wild-type alleles of Rht-B1 and Rht-D1 as independent determinants of thousand-grain weight and kernel number per spike in wheat. Mol Breed 32:771–783
Zhang N, Fan XL, Cui F et al (2017) Characterization of the temporal and spatial expression of wheat (Triticum aestivum L.) plant height at the QTL level and their influence on yield-related traits. Theor Appl Genet 130:1235–1252
Zhang JL, Gizaw SA, Bossolini E et al (2018a) Identification and validation of QTL for grain yield and plant water status under contrasting water treatments in fall-sown spring wheats. Theor Appl Genet 131:1741–1759
Zhang B, Wang XG, Wang XL et al (2018b) Molecular characterization of a novel vernalization allele Vrn-B1d and its effect on heading time in Chinese wheat (Triticum aestivum L.) landrace Hongchunmai. Mol Breed 38:127
Acknowledgements
This work was supported by the Transgenic Special Item of China (2016ZX08002003-002 and 2016ZX08009-003), the National Natural Science Foundation of China (31520103911, 31871610, 31901491 and 31901492), and the Agricultural Variety Improvement Project of Shandong Province (2019LZGC010).
Author information
Authors and Affiliations
Contributions
LK and HW designed this research. JH carried out the research and then wrote the first draft of the manuscript. XW conducted the data analysis and revised this paper. GZ, WC, YH, XM, and SX participated in the field experiments and data collection. PJ constructed the integrated linkage map. JJ provided the materials. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare that they have no competing interests.
Ethical standards
I declare on behalf of my co-authors that the work described is original, previously unpublished research, and not under consideration for publication elsewhere. The experiments in this study comply with the current laws of China.
Additional information
Communicated by Gary Muehlbauer.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hu, J., Wang, X., Zhang, G. et al. QTL mapping for yield-related traits in wheat based on four RIL populations. Theor Appl Genet 133, 917–933 (2020). https://doi.org/10.1007/s00122-019-03515-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03515-w