Abstract
Non-linear kernel methods can be approximated by fast linear ones using suitable explicit feature maps allowing their application to large scale problems. We investigate how convolution kernels for structured data are composed from base kernels and construct corresponding feature maps. On this basis we propose exact and approximative feature maps for widely used graph kernels based on the kernel trick. We analyze for which kernels and graph properties computation by explicit feature maps is feasible and actually more efficient. In particular, we derive approximative, explicit feature maps for state-of-the-art kernels supporting real-valued attributes including the GraphHopper and graph invariant kernels. In extensive experiments we show that our approaches often achieve a classification accuracy close to the exact methods based on the kernel trick, but require only a fraction of their running time. Moreover, we propose and analyze algorithms for computing random walk, shortest-path and subgraph matching kernels by explicit and implicit feature maps. Our theoretical results are confirmed experimentally by observing a phase transition when comparing running time with respect to label diversity, walk lengths and subgraph size, respectively.
Similar content being viewed by others
1 Introduction
Analyzing complex data is becoming more and more important. In numerous application domains, e.g., chem- and bioinformatics, neuroscience, or image and social network analysis, the data is structured and hence can naturally be represented as graphs. To achieve successful learning we need to exploit the rich information inherent in the graph structure and the annotations of vertices and edges. A popular approach to mining structured data is to design graph kernels measuring the similarity between pairs of graphs. The graph kernel can then be plugged into a kernel machine, such as support vector machine or Gaussian process, for efficient learning and prediction.
The kernel-based approach to predictive graph mining requires a positive semidefinite (p.s.d.) kernel function between graphs. Graphs, composed of labeled vertices and edges, possibly enriched with continuous attributes, however, are not fixed-length vectors but rather complicated data structures, and thus standard kernels cannot be used. Instead, the general strategy to design graph kernels is to decompose graphs into small substructures among which kernels are defined following the concept of convolution kernels due to Haussler (1999). The graph kernel itself is then a combination of the kernels between the possibly overlapping parts. Hence the various graph kernels proposed in the literature mainly differ in the way the parts are constructed and in the similarity measure used to compare them. Moreover, existing graph kernels differ in their ability to exploit annotations, which may be categorical labels or real-valued attributes on the vertices and edges.
We recall basic facts on kernels, which have decisive implications on computational aspects. A kernel on a non-empty set \(\mathcal {X} \) is a positive semidefinite function \(k :\mathcal {X} \times \mathcal {X} \rightarrow \mathbb {R} \). Equivalently, a function k is a kernel if there is a feature map\(\phi :\mathcal {X} \rightarrow \mathcal {H} \) to a Hilbert space \(\mathcal {H}\) with inner product \(\langle \cdot , \cdot \rangle \), such that \(k(x,y) = \langle \phi (x),\phi (y) \rangle \) for all x and y in \(\mathcal {X} \). This equivalence yields two algorithmic strategies to compute kernels on graphs:
-
(i)
One way is functional computation, e.g., from closed-form expressions. In this case the feature map is not necessarily known and the feature space may be of infinite dimension. Therefore, we refer to this approach based on the famous kernel trick as implicit computation.
-
(ii)
The other strategy is to compute the feature map \(\phi (G)\) for each graph Gexplicitly to obtain the kernel values from the dot product between pairs of feature vectors. These feature vectors commonly count how often certain substructures occur in a graph.
The used strategy has a crucial effect on the running time of the kernel method at the higher level. Kernel methods supporting implicit kernel computation are often slower than linear ones based on explicit feature maps assuming that the feature vectors are of a manageable size. The running time for training support vector machines, for example, is linear in the training set size when assuming that the feature vectors have a constant number of non-zero components (Joachims 2006). For this reason, approximative explicit feature maps of various popular kernels for vectorial data have been studied extensively (Rahimi and Recht 2008; Vedaldi and Zisserman 2012). This, however, is not the case for graph kernels, which are typically proposed using one method of computation, either implicit or explicit. Graph kernels using explicit feature maps essentially transform graphs into vectorial data in a preprocessing step. These kernels are scalable, but are often restricted to graphs with discrete labels. Unique advantages of the implicit computation are that (i) kernels for composed objects can be obtained by combining established kernels on their parts exploiting well-known closure properties of kernels; (ii) the number of possible features may be high—in theory infinite—while the function remains polynomial-time computable.
Previously proposed graph kernels that are computed implicitly typically support specifying arbitrary kernels for vertex annotations, but do not scale to large graphs and data sets. Even when approximative explicit feature maps of the kernel on vertex annotations are known, it is not clear how to obtain (approximative) feature maps for the graph kernel.
Our contribution We study under which conditions the computation of an explicit mapping from graphs to a finite-dimensional feature spaces is feasible and efficient. To achieve our goal, we discuss feature maps corresponding to closure properties of kernels and general convolution kernels with a focus on the size and sparsity of their feature vectors. Our theoretical analysis identifies a trade-off between running time and flexibility.
Building on the systematic construction of feature maps we obtain new algorithms for explicit graph kernel computation, which allow to incorporate (approximative) explicit feature maps of kernels on vertex annotations. Thereby known approximation results for kernels on continuous data are lifted to kernels for graphs with continuous annotations. More precisely, we introduce the class of weighted vertex kernels and show that it generalizes state-of-the-art kernels for graphs with continuous attributes, namely the GraphHopper kernel (Feragen et al. 2013) and an instance of the graph invariant kernels (Orsini et al. 2015). We derive explicit feature maps with approximation guarantees based on approximative feature maps of the base kernels to compare annotations. Then, we propose and analyze algorithms for computing fixed length walk kernels by explicit and implicit feature maps. We investigate shortest-path kernels (Borgwardt and Kriegel 2005) and subgraph matching kernels (Kriege and Mutzel 2012) and put the related work into the context of our systematic study. Given this, we are finally able to experimentally compare the running times of both computation strategies systematically with respect to the label diversity, data set size, and substructure size, i.e., walk length and subgraph size. As it turns out, there exists a computational phase transition for walk and subgraph kernels. Our experimental results for weighted vertex kernels show that their computation by explicit feature maps is feasible and provides a viable alternative even when comparing graphs with continuous attributes.
Extension of the conference paper The present paper is a significant extension of a previously published conference paper (Kriege et al. 2014). In the following we list the main contributions that were not included in the conference version.
-
Feature maps of composed kernels We review closure properties of kernels, the corresponding feature maps and the size and sparsity of the feature vectors. Based on this, we obtain explicit feature maps for convolution kernels with arbitrary base kernels. This generalizes the result of the conference paper, where binary base kernel were considered.
-
Weighted vertex kernels We introduce weighted vertex kernels for attributed graphs, which generalize the GraphHopper kernel (Feragen et al. 2013) and graph invariant kernels (Orsini et al. 2015). Weighted vertex kernels were not considered in the conference paper.
-
Construction of explicit feature maps We derive explicit feature maps for weighted vertex kernels and the shortest-path kernel (Borgwardt and Kriegel 2005) supporting base kernels with explicit feature maps for the comparison of attributes. We prove approximation guarantees in case of approximative feature maps of base kernels. This contribution is not contained in the conference paper, where only the explicit computation of the shortest-path kernel for graphs with discrete labels was discussed.
-
Fixed length walk kernels We generalize the explicit computation scheme to support arbitrary vertex and edge kernels with explicit feature maps for the comparison of attributes. In the conference paper only binary kernels were considered. Moreover, we have significantly expanded the section on walk kernels by spelling out all proofs, adding illustrative figures and clarifying the relation to the k-step random walk kernel as defined by Sugiyama and Borgwardt (2015).
-
Experimental evaluation We largely extended our evaluation, which now includes experiments for the novel computation schemes of graph kernels as well as a comparison between a graphlet kernel and the subgraph matching kernel (Kriege and Mutzel 2012).
Outline In Sect. 2 we discuss the related work and proceed by fixing the notation in Sect. 3. In Sect. 4 we review closure properties of kernels and the corresponding feature maps. Moreover, we derive feature maps for general convolution kernels. In Sect. 5 we propose algorithms for computing graph kernels and systematically construct explicit feature maps building on these results. We introduce weighted vertex kernels and derive (approximative) explicit feature maps. We discuss the fixed length walk kernel and propose algorithms for explicit and implicit computation. Moreover, we discuss the shortest-path graph kernel as well as the graphlet and subgraph matching kernel regarding explicit and implicit computation. Section 6 presents the results of our experimental evaluation.
2 Related work
In the following we review existing kernels based on explicit or implicit computation and discuss embedding techniques for attributed graphs. We focus on the approaches most relevant for our work and refer the reader to the survey articles (Vishwanathan et al. 2010; Ghosh et al. 2018; Zhang et al. 2018b; Kriege 2019) for a more comprehensive overview.
2.1 Graph kernels
Most graph kernels decompose graphs into substructures and count their occurrences to obtain a feature vector. The kernel function then counts the co-occurrences of features in two graphs by taking the dot product between their feature vectors. The graphlet kernel, for example, counts induced subgraphs of size \(k \in \{3,4,5\}\) of unlabeled graphs according to \(K(G,H) = {\mathbf {f}}_{G}^\top {\mathbf {f}}_{H}\), where \({\mathbf {f}}_{G}\) and \({\mathbf {f}}_{H}\) are the subgraph feature vectors of G and H, respectively (Shervashidze et al. 2009). The cyclic pattern kernel is based on cycles and trees and maps the graphs to substructure indicator features, which are independent of the substructure frequency (Horváth et al. 2004). The Weisfeiler-Lehman subtree kernel counts label-based subtree patterns according to \(K_d(G, H) = \sum _{i = 1}^h K(G_i, H_i)\), where \(K(G_i, H_i) = \langle {\mathbf {f}}^{(i)}_{G}{\mathbf {f}}^{(i)}_{H} \rangle \) and \({\mathbf {f}}^{(i)}_{G}\) is a feature vector counting subtree-patterns in G of depth i (Shervashidze et al. 2011; Shervashidze and Borgwardt (2009)). A subtree-pattern is a tree rooted at a particular vertex where each level contains the neighbors of its parent vertex; the same vertices can appear repeatedly. Other graph kernels on subtree-patterns have been proposed in the literature, e.g., Ramon and Gärtner (2003), Harchaoui and Bach (2007), Bai et al. (2015) and Hido and Kashima (2009). In a similar spirit, the propagation kernel iteratively counts similar label or attribute distributions to create an explicit feature map for efficient kernel computation (Neumann et al. 2016). Martino et al. (2012) proposed to decompose graphs into multisets of ordered directed acyclic graphs, which are compared by extended tree kernels. While convolution kernels decompose graphs into their parts and sum over all pairs, assignment kernels are obtained from an optimal bijection between parts (Fröhlich et al. 2005). Since this does not lead to valid kernels in general (Vert 2008; Vishwanathan et al. 2010), various approaches to overcome this obstacle have been developed (Johansson and Dubhashi 2015; Schiavinato et al. 2015; Kriege et al. 2016; Nikolentzos et al. 2017). Several kernels have been proposed with the goal to take graph structure at different scales into account, e.g., using k-core decomposition (Nikolentzos et al. 2018) or spectral properties (Kondor and Pan 2016). Yanardag and Vishwanathan (2015) combine neural techniques from language modeling with state-of-the-art graph kernels in order to incorporate similarities between the individual substructures. Such similarities were specifically designed for the substrutures used by the graphlet and the Weisfeiler-Lehman subtree kernel, among others. Narayanan et al. (2016) discuss several problems of the proposed approach to obtain substructure similarities and introduce subgraph2vec to overcome these issues.
Many real-world graphs have continuous attributes such as real-valued vectors attached to their vertices and edges. For example, the vertices of a molecular graph may be annotated by the physical and chemical properties of the atoms they represent. The kernels based on counting co-occurrences described above, however, consider two substructures as identical if they match exactly, structure-wise as well as attribute-wise, and as completely different otherwise. For attributed graphs it is desirable to compare annotations by more complex similarity measures such as the Gaussian RBF kernel. The kernels discussed in the following allow user-defined kernels for the comparison of vertex and edge attributes. Moreover, they compare graphs in a way that takes the interplay between structure and attributes into account and are therefore suitable for graphs with continuous attributes.
Random walk kernels add up a score for all pairs of walks that two graphs have in common, whereas vertex and edge attributes encountered on walks can be compared by user-specified kernels. For random walk kernels implicit computation schemes based on product graphs have been proposed. The product graph \(G_{\times }\) has a vertex for each pair of vertices in the original graphs. Two vertices in the product graph are neighbors if the corresponding vertices in the original graphs are both neighbors as well. Product graphs have some nice properties making them suitable for the computation of graph kernels. First, the adjacency matrix \(A_{\times }\) of a product graph is the Kronecker product of the adjacency matrices A and \(A'\) of the original graphs, i.e., \(A_{\times } = A \otimes A'\), same holds for the weight matrix \(W_{\times }\) when employing an edge kernel. Further, there is a one-to-one correspondence between walks on the product graph and simultaneous walks on the original graphs (Gärtner et al. 2003). The random walk kernel introduced by Vishwanathan et al. (2010) is now given by
where \(p_\times \) and \(q_\times \) are starting and stopping probability distributions and \(\mu _l\) coefficients such that the sum converges. Several variations of the random walk kernel have been introduced in the literature. The geometric random walk kernel originally introduced by Gärtner et al. (2003) counts walks with the same sequence of discrete labels and is a predecessor of the general formulation presented above. The description of the random walk kernel by Kashima et al. (2003) is motivated by a probabilistic view on kernels and based on the idea of so-called marginalized kernels. The method was extended to avoid tottering and the efficiency was improved by label refinement (Mahé et al. 2004). Several methods for computing Eq. (1) were proposed by Vishwanathan et al. (2010) achieving different running times depending on a parameter k, which is the number of fixed-point iterations, power iterations and the effective rank of \(W_\times \), respectively. The running times to compare graphs with n vertices also depend on the edge labels of the input graphs and the desired edge kernel. For unlabeled graphs the running time \(\mathcal {O}(n^3)\) is achieved and \(\mathcal {O}(dkn^3)\) for labeled graphs, where d is the size of the label alphabet. The same running time is obtained for edge kernels with a d-dimensional feature space, while \(\mathcal {O}(kn^4)\) time is required in the infinite case. For sparse graphs \(\mathcal {O}(kn^2)\) is obtained in all cases. Further improvements of the running time were subsequently obtained by non-exact algorithms based on low rank approximations (Kang et al. 2012). These random walk kernels take all walks without a bound on length into account. However, in several applications it has been reported that only walks up to a certain length have been considered, e.g., for the prediction of protein functions (Borgwardt et al. 2005) or image classification (Harchaoui and Bach 2007). This might suggest that it is not necessary or even not beneficial to consider the infinite number of possible walks to obtain a satisfying prediction accuracy. Moreover, the phenomenon of halting in random walk kernels has been studied recently (Sugiyama and Borgwardt 2015), which refers to the fact that long walks are down-weighted such that the kernel is in fact dominated by walks of length 1.
Another substructure used to measure the similarity among graphs are shortest paths. Borgwardt and Kriegel (2005) proposed the shortest-path kernel, which compares two graphs based on vertex pairs with similar shortest-path lengths. The GraphHopper kernel compares the vertices encountered while hopping along shortest paths by a user-specified kernel (Feragen et al. 2013). Similar to the graphlet kernel, the subgraph matching kernel compares subgraphs of small size, but allows to score mappings between them according to vertex and edge kernels (Kriege and Mutzel 2012). Further kernels designed specifically for graphs with continuous attributes exist (Orsini et al. 2015; Su et al. 2016; Martino et al. 2018).
2.2 Embedding techniques for attributed graphs
Kernels for attributed graphs often allow to specify arbitrary kernels for comparing attributes and are computed using the kernel trick without generating feature vectors. Moreover, several approaches for computing vector representations for attributed graphs have been proposed. These, however, do not allow specifying a function for comparing attributes. The similarity measure that is implicitly used to compare attributes is typically not known. This is the case for recent deep learning approaches as well as for some kernels proposed for attributed graphs.
Deep learning on graphs Recently, a number of approaches to graph classification based upon neural networks have been proposed. Here a vectorial representation for each vertex is learned iteratively from the vertex annotations of its neighbors using a parameterized (differentiable) neighborhood aggregation function. Eventually, the vector representations for the individual vertices are combined to obtain a vector representation for the graph, e.g., by summation.
The parameters of the aggregation function are learned together with the parameters of the classification or regression algorithm, e.g., a neural network. More refined approaches use differential pooling operators based on sorting (Zhang et al. 2018a) and soft assignments (Ying et al. 2018b). Most of these neural approaches fit into the framework proposed by Gilmer et al. (2017). Notable instances of this model include neural fingerprints (Duvenaud et al. 2015), GraphSAGE (Hamilton et al. 2017a), and the spectral approaches proposed by Bruna et al. (2014), Defferrard et al. (2016) and Kipf and Welling (2017)—all of which descend from early work, see, e.g., Merkwirth and Lengauer (2005) and Scarselli et al. (2009).
These methods show promising results on several graph classification benchmarks, see, e.g., Ying et al. (2018b), as well as in applications such as protein–protein interaction prediction (Fout et al. 2017), recommender systems (Ying et al. 2018a), and the analysis of quantum interactions in molecules (Schütt et al. 2017). A survey of recent advancements can be found in Hamilton et al. (2017b). With these approaches, the vertex attributes are aggregated for each graph and not directly compared between the graphs. Therefore, it is not obvious how the similarity of vertex attributes is measured.
Explicit feature maps of kernels for attributed graphs Graph kernels supporting complex annotations typically use implicit computation schemes and do not scale well. Whereas graphs with discrete labels are efficiently compared by graph kernels based on explicit feature maps. Kernels limited to graphs with categorical labels can be applied to attributed graphs by discretization of the continuous attributes, see, e.g., Neumann et al. (2016). Morris et al. (2016) proposed the hash graph kernel framework to obtain efficient kernels for graphs with continuous labels from those proposed for discrete ones. The idea is to iteratively turn continuous attributes into discrete labels using randomized hash functions. A drawback of the approach is that so-called independent k-hash families must be known to guarantee that the approach approximates attribute comparisons by the kernel k. In practice locality-sensitive hashing is used, which does not provide this guarantee, but still achieves promising results. To the best of our knowledge no results on explicit feature maps of kernels for graphs with continuous attributes that are compared by a well-defined similarity measure such as the Gaussian RBF kernel are known.
However, explicit feature maps of kernels for vectorial data have been studied extensively. Starting with the seminal work by Rahimi and Recht (2008), explicit feature maps of various popular kernels have been proposed, cf. (Vedaldi and Zisserman 2012; Kar and Karnick 2012; Pham and Pagh 2013, and references therein). In this paper, we build on this line of work to obtain kernels for graphs, where individual vertices and edges are annotated by vectorial data. In contrast to the hash graph kernel framework our goal is to lift the known approximation results for kernels on vectorial data to kernels for graphs annotated with vectorial data.
3 Preliminaries
An (undirected) graphG is a pair (V, E) with a finite set of verticesV and a set of edges\(E \subseteq \{ \{u,v\} \subseteq V \mid u \ne v \}\). We denote the set of vertices and the set of edges of G by V(G) and E(G), respectively. For ease of notation we denote the edge \(\{u,v\}\) in E(G) by uv or vu and the set of all graphs by \(\mathcal {G}\). A graph \(G' = (V',E')\) is a subgraph of a graph \(G=(V,E)\) if \(V' \subseteq V\) and \(E' \subseteq E\). The subgraph \(G'\) is said to be induced if \(E' = \{uv \in E \mid u,v \in V' \}\) and we write \(G' \subseteq G\). We denote the neighborhood of a vertex v in V(G) by \({{\,\mathrm{N}\,}}(v) = \{ u \in V(G) \mid vu \in E(G) \}\).
A labeled graph is a graph G endowed with an label function\(\tau :V(G) \rightarrow \varSigma \), where \(\varSigma \) is a finite alphabet. We say that \(\tau (v)\) is the label of v for v in V(G). An attributed graph is a graph G endowed with a function \(\tau :V(G) \rightarrow \mathbb {R} ^d\), \(d \in \mathbb {N}^{+}\), and we say that \(\tau (v)\) is the attribute of v. We denote the base kernel for comparing vertex labels and attributes by \(k_V\) and, for short, write \(k_V(u,v)\) instead of \(k_V(\tau (u),\tau (v))\). The above definitions directly extend to graphs, where edges have labels or attributes and we denote the base kernel by \(k_E\). We refer to \(k_V\) and \(k_E\) as vertex kernel and edge kernel, respectively, and assume both to take non-negative values only.
Let \({\mathsf {T}}_k\) be the running time for evaluating a kernel for a pair of graphs, \({\mathsf {T}}_\phi \) for computing a feature vector for a single graph and \({\mathsf {T}}_{\text {dot}}\) for computing the dot product between two feature vectors. Computing an \(n \times n\) matrix with all pairwise kernel values for n graphs requires (i) time \(\mathcal {O}(n^2 {\mathsf {T}}_k)\) using implicit feature maps, and (ii) time \(\mathcal {O}(n {\mathsf {T}}_{\phi } + n^2 {\mathsf {T}}_{\text {dot}})\) using explicit feature maps. Clearly, explicit computation can only be competitive with implicit computation, when the time \({\mathsf {T}}_{\text {dot}}\) is smaller than \({\mathsf {T}}_k\). In this case, however, even a time-consuming feature mapping \({\mathsf {T}}_\phi \) pays off with increasing data set size. The running time \({\mathsf {T}}_{\text {dot}}\) depends on the data structure used to store feature vectors. Since feature vectors for graph kernels often contain many components that are zero, we consider sparse data structures, which expose running times depending on the number of non-zero components instead of the actual number of all components. For a vector v in \(\mathbb {R} ^d\), we denote by \(\mathsf {nz}(v)\) the set of indices of the non-zero components of v and let \(\mathsf {nnz}(v) = |\mathsf {nz}(v)|\) the number of non-zero components. Using hash tables the dot product between \(\varPhi _1\) and \(\varPhi _2\) can be realized in time \({\mathsf {T}}_{\text {dot}}=\mathcal {O}(\min \{\mathsf {nnz}(\varPhi _1), \mathsf {nnz}(\varPhi _2)\})\) in the average case.
4 Basic kernels, composed kernels and their feature maps
Graph kernels, in particular those supporting user-specified kernels for annotations, typically employ closure properties. This allows to decompose graphs into parts that are eventually the annotated vertices and edges. The graph kernel then is composed of base kernels applied to the annotations and annotated substructures, respectively. We first consider the explicit feature maps of basic kernels and then review closure properties of kernels and discuss how to obtain their explicit feature maps. The results are summarized in Table 1. This forms the basis for the systematic construction of explicit feature maps of graph kernels according to their composition of base kernels later in Sect. 5.
Some of the basic results on the construction of feature maps and their detailed proofs can be found in the text book by Shawe-Taylor and Cristianini (2004). Going beyond that, we discuss the sparsity of the obtained feature vectors in detail. This has an essential impact on the efficiency in practice, when sparse data structures are used and a large number of the components of a feature vector is zero. Indeed the running times we observed experimentally in Sect. 6 can only be explained taking the sparsity into account.
4.1 Dirac and binary kernels
We discuss feature maps for basic kernels often used for the construction of kernels on structured objects. The Dirac kernel \(k_\delta \) on \(\mathcal {X} \) is defined by \(k_\delta (x,y) = 1\), if \(x=y\) and 0 otherwise. For \(\mathcal {X}\) a finite set, it is well-known that \(\phi :\mathcal {X} \rightarrow \{0,1\}^{|\mathcal {X} |}\) with components indexed by \(i \in \mathcal {X} \) and defined as \(\phi (x)_i = 1\) if \(i=x\), and 0 otherwise, is a feature map of the Dirac kernel.
The requirement that two objects are equal is often too strict. When considering two subgraphs, for example, the kernel should take the value 1 if the graphs are isomorphic and 0 otherwise. Likewise, two vertex sequences corresponding to walks in graphs should be regarded as identical if their vertices have the same labels. We discuss this more general concept of kernels and their properties in the following. We say a kernel k on \(\mathcal {X} \) is binary if k(x, y) is either 0 or 1 for all \(x,y \in \mathcal {X} \). Given a binary kernel, we refer to
as the relation on \(\mathcal {X} \)induced byk. Next we will establish several properties of this relation, which will turn out to be useful for the construction of a feature map.
Lemma 1
Let k be a binary kernel on \(\mathcal {X} \), then \(x \sim _k y \Longrightarrow x \sim _k x\) holds for all \(x,y \in \mathcal {X} \).
Proof
Assume there are \(x,y \in \mathcal {X} \) such that \(x \not \sim _k x\) and \(x \sim _k y\). By the definition of \(\sim _k\) we obtain \(k(x,x)=0\) and \(k(x,y)=1\). The symmetric kernel matrix obtained by k for \(X=\{x,y\}\) thus is either \(\bigl ({\begin{matrix} 0&{}1\\ 1&{}0 \end{matrix}} \bigr )\) or \(\bigl ({\begin{matrix} 0&{}1\\ 1&{}1 \end{matrix}} \bigr )\), where we assume that the first row and column is associated with x. Both matrices are not p.s.d. and, thus, k is not a kernel contradicting the assumption. \(\square \)
Lemma 2
Let k be a binary kernel on \(\mathcal {X} \), then \(\sim _k\) is a partial equivalence relation meaning that the relation \(\sim _k\) is (i) symmetric, and (ii) transitive.
Proof
Property (i) follows from the fact that k must be symmetric according to definition. Assume property (ii) does not hold. Then there are \(x,y,z \in \mathcal {X} \) with \(x \sim _k y \wedge y \sim _k z\) and \(x \not \sim _k z\). Since \(x \ne z\) must hold according to Lemma 1 we can conclude that \(X=\{x,y,z\}\) are pairwise distinct. We consider a kernel matrix \(\mathbf {K}\) obtained by k for X and assume that the first, second and third row as well as column is associated with x, y and z, respectively. There must be entries \(k_{12}=k_{21}=k_{23}=k_{32}=1\) and \(k_{13}=k_{31}=0\). According to Lemma 1 the entries of the main diagonal \(k_{11}=k_{22}=k_{33}=1\) follow. Consider the coefficient vector \(\mathbf {c}\) with \(c_1=c_3=1\) and \(c_2=-1\), we obtain \(\mathbf {c}^\top \mathbf {K} \mathbf {c} = -1.\) Hence, \(\mathbf {K}\) is not p.s.d. and k is not a kernel contradicting the assumption. \(\square \)
We use these results to construct a feature map for a binary kernel. We restrict our consideration to the set \(\mathcal {X} _{{{\,\mathrm{ref}\,}}} = \{ x \in \mathcal {X} \mid x \sim _k x \}\), on which \(\sim _k\) is an equivalence relation. The quotient set \(\mathcal {Q}_k=\mathcal {X} _{{{\,\mathrm{ref}\,}}}/\!\!\sim _k\) is the set of equivalence classes induced by \(\sim _k\). Let \([x]_k\) denote the equivalence class of \(x \in \mathcal {X} _{{{\,\mathrm{ref}\,}}}\) under the relation \(\sim _k\). Let \(k_\delta \) be the Dirac kernel on the equivalence classes \(\mathcal {Q}_k\), then \(k(x,y) = k_\delta ([x]_k,[y]_k)\) and we obtain the following result.
Proposition 1
Let k be a binary kernel with \(\mathcal {Q}_k = \{Q_1,\dots ,Q_d\}\), then \(\phi :\mathcal {X} \rightarrow \{0,1\}^{d}\) with \(\phi (x)_i = 1\) if \(Q_i=[x]_k\), and 0 otherwise, is a feature map of k.
4.2 Closure properties
For a kernel k on a non-empty set \(\mathcal {X} \) the function \(k^{\alpha }(x,y) = \alpha k(x,y)\) with \(\alpha \) in \(\mathbb {R} _{\ge 0}\) is again a kernel on \(\mathcal {X} \). Let \(\phi \) be a feature map of k, then \(\phi ^{\alpha }(x) = \sqrt{\alpha }\phi (x)\) is a feature map of \(k^{\alpha }\). For addition and multiplication, we get the following result.
Proposition 2
(Shawe-Taylor and Cristianini 2004, pp. 75 sqq.)
Let \(k_1, \dots , k_D\) for \(D>0\) be kernels on \(\mathcal {X} \) with feature maps \(\phi _1, \dots , \phi _D\) of dimension \(d_1, \dots , d_D\), respectively. Then
are again kernels on \(\mathcal {X} \). Moreover,
are feature maps for \(k^{+}\) and \(k^{\bullet }\) of dimension \(\sum ^D_{i=1} d_i\) and \(\prod ^D_{i=1} d_i\), respectively. Here \(\oplus \) denotes the concatenation of vectors and \(\otimes \) the Kronecker product.
Remark 1
In case of \(k_1 = k_2 = \dots = k_D\), we have \(k^{+}(x,y)=D k_1(x,y)\) and a \(d_1\)-dimensional feature map can be obtained. For \(k^{\bullet }\) we have \(k_1(x,y)^D\), which yet does not allow for a feature space of dimension smaller than \(d_1^D\) in general.
We state an immediate consequence of Proposition 2 regarding the sparsity of the obtained feature vectors explicitly.
Corollary 1
Let \(k_1, \dots , k_D\) and \(\phi _1, \dots , \phi _D\) be defined as above, then
4.3 Kernels on sets
In the following we derive an explicit mapping for kernels on finite sets. This result will be needed in the succeeding section for constructing an explicit feature map for the R-convolution kernel. Let \(\kappa \) be a base kernel on a set U, and let X and Y be finite subsets of U. Then the cross product kernel or derived subset kernel on \(\mathcal {P}(U)\) is defined as
Let \(\phi \) be a feature map of \(\kappa \), then the function
is a feature map of the cross product kernel (Shawe-Taylor and Cristianini 2004, Proposition 9.42). In particular, the feature space of the cross product kernel corresponds to the feature space of the base kernel; both have the same dimension. For \(\kappa =k_\delta \) the Dirac kernel \(\phi ^\times (X)\) maps the set X to its characteristic vector, which has |U| components and |X| non-zero elements. When \(\kappa \) is a binary kernel as discussed in Sect. 4 the number of components reduces to the number of equivalence classes of \(\sim _\kappa \) and the number of non-zero elements becomes the number of cells in the quotient set \(X/\!\!\sim _\kappa \). In general, we obtain the following result as an immediate consequence of Eq. (3).
Corollary 2
Let \(\phi ^{\times }\) be the feature map of the cross product kernel and \(\phi \) the feature map of its base kernel, then
A crucial observation is that the number of non-zero components of a feature vector depends on both, the cardinality and structure of the set X and the feature map \(\phi \) acting on the elements of X. It is as large as possible when each element of X is mapped by \(\phi \) to a feature vector with distinct non-zero components.
4.4 Convolution kernels
Haussler (1999) proposed R-convolution kernels as a generic framework to define kernels between composite objects. In the following we derive feature maps for such kernels by using the basic closure properties introduced in the previous sections. Thereby, we generalize the result presented in (Kriege et al. 2014).
Definition 1
Suppose \(x \in \mathcal {R} = \mathcal {R}_1 \times \cdots \times \mathcal {R}_n\) are the parts of \(X \in \mathcal {X} \) according to some decomposition. Let \(R \subseteq \mathcal {X} \times \mathcal {R}\) be a relation such that \((X,x) \in R\) if and only if X can be decomposed into the parts x. Let \(R(X) = \{ x \mid (X, x) \in R \}\) and assume R(X) is finite for all \(X \in \mathcal {X} \). The R-convolution kernel is
where \(\kappa _i\) is a kernel on \(\mathcal {R}_i\) for all \(i \in \{1,\dots ,n\}\).
Assume that we have explicit feature maps for the kernels \(\kappa _i\). We first note that a feature map for \(\kappa \) can be obtained from the feature maps for \(\kappa _i\) by Proposition 2.Footnote 1 In fact, Eq. (4) for arbitrary n can be obtained from the case \(n=1\) for an appropriate choice of \(\mathcal {R}_1\) and \(k_1\) as noted by Shin and Kuboyama (2010). If we assume \(\mathcal {R} = \mathcal {R}_1 = U\), the R-convolution kernel boils down to the crossproduct kernel and we have \(k^\star (X,Y) = k^\times (R(X),R(Y))\), where both employ the same base kernel \(\kappa \). We use this approach to develop explicit mapping schemes for graph kernels in the following. Let \(\phi \) be a feature map for \(\kappa \) of dimension d, then from Eq. (3), we obtain an explicit mapping of dimension d for the R-convolution kernel according to
As discussed in Sect. 4.3 the sparsity of \(\phi ^{\star }(X)\) simultaneous depends on the number of parts and their relation in the feature space of \(\kappa \).
Kriege et al. (2014) considered the special case that \(\kappa \) is a binary kernel, cf. Sect. 4.1. From Proposition 1 and Eq. (5) we directly obtain their result as special case.
Corollary 3
(Kriege et al. (2014), Theorem 3) Let \(k^\star \) be an R-convolution kernel with binary kernel \(\kappa \) and \(\mathcal {Q}_\kappa = \{Q_1,\dots ,Q_d\}\), then \(\phi ^\star :\mathcal {X} \rightarrow \mathbb {N} ^d\) with \(\phi (x)_i = |Q_i \cap X|\) is a feature map of \(k^\star \).
5 Computing graph kernels by explicit and implicit feature maps
Building on the systematic construction of feature maps of kernels, we discuss explicit and implicit computation schemes of graph kernels. We first introduce weighted vertex kernels. This family of kernels generalizes the GraphHopper kernel (Feragen et al. 2013) and graph invariant kernels (Orsini et al. 2015) for attributed graphs, which were recently proposed with an implicit method of computation. We derive (approximative) explicit feature maps for weighted vertex kernels. Then, we develop explicit and implicit methods of computation for fixed length walk kernels, which both exploit sparsity for efficient computation. Finally, we discuss shortest-path and subgraph kernels for which both computation schemes have been considered previously and put them in the context of our systematic study. We empirically study both computation schemes for graph kernels confirming our theoretical results experimentally in Sect. 6.
5.1 Weighted vertex kernels
Kernels suitable for attributed graphs typically use user-defined kernels for the comparison of vertex and edge annotations such as real-valued vectors. The graph kernel is then obtained by combining these kernels according to closure properties. Recently proposed kernels for attributed graphs such as GraphHopper (Feragen et al. 2013) and graph invariant kernels (Orsini et al. 2015) use separate kernel functions for the graph structure and vertex annotations. They can be expressed as
where \(k_V\) is a user-specified kernel comparing vertex attributes and \(k_W\) is a kernel that determines a weight for a vertex pair based on the individual graph structures. Hence, in the following we refer to Eq. (6) as weighted vertex kernel. Kernels belonging to this family are easily identifiable as instances of R-convolution kernels, cf. Definition 1.
5.1.1 Weight kernels
We discuss two kernels for attributed graphs, which have been proposed recently and can bee seen as instances of weighted vertex kernels.
Graph invariant kernels One approach to obtain weights for pairs of vertices is to compare their neighborhood by the classical Weisfeiler-Lehman label refinement (Shervashidze et al. 2011; Orsini et al. 2015). For a parameter h and a graph G with uniform initial labels \(\tau _0\), a sequence \((\tau _1,\dots ,\tau _h)\) of refined labels referred to as colors is computed, where \(\tau _i\) is obtained from \(\tau _{i-1}\) by the following procedure. Sort the multiset of colors \({\lbrace \lbrace }\tau _{i-1}(u) \mid \, vu \in E(G){\rbrace \rbrace }\) for every vertex v to obtain a unique sequence of colors and add \(\tau _{i-1}(v)\) as first element. Assign a new color \(\tau _i(v)\) to every vertex v by employing an injective mapping from color sequences to new colors. A reasonable implementation of \(k_W\) motivated along the lines of graph invariant kernels (Orsini et al. 2015) is
where \(\tau _i(v)\) denotes the discrete label of the vertex v after the ith iteration of Weisfeiler-Lehman label refinement of the underlying unlabeled graph. Intuitively, this kernel reflects to what extent the two vertices have a structurally similar neighborhood.
GraphHopper kernel Another graph kernel, which fits into the framework of weighted vertex kernels, is the GraphHopper kernel (Feragen et al. 2013) with
Here \(\mathbf {M}(v)\) and \(\mathbf {M}(v')\) are \(\delta \times \delta \) matrices, where the entry \(\mathbf {M}(v)_{ij}\) for v in V(G) counts the number of times the vertex v appears as the ith vertex on a shortest path of discrete length j in G, where \(\delta \) denotes the maximum diameter over all graphs, and \(\langle \cdot , \cdot \rangle _F\) is the Frobenius inner product.
5.1.2 Vertex kernels
For graphs with multi-dimensional real-valued vertex attributes in \(\mathbb {R} ^d\) one could set \(k_V\) to the Gaussian RBF kernel \(k_{\text {RBF}}\) or the dimension-wise product of the hat kernel \(k_{\varDelta }\), respectively, i.e.,
Here, \(\sigma \) and \(\delta \) are parameters controlling the decrease of the kernel value with increasing discrepancy between the two input data points.
5.1.3 Computing explicit feature maps
In the following we derive an explicit mapping for weighted vertex kernels. Notice that Eq. (6) is an instance of Definition 1. Hence, by Proposition 2 and Eq. (5), we obtain an explicit mapping \(\phi ^\text {WV}\) of weighted vertex kernels.
Proposition 3
Let \(K^\text {WV}\) be a weighted vertex kernel according to Eq. (6) with \(\phi ^W\) and \(\phi ^V\) feature maps for \(k_W\) and \(k_V\), respectively. Then
is a feature map for \(K^\text {WV}\).
Widely used kernels for the comparison of attributes, such as the Gaussian RBF kernel, do not have feature maps of finite dimension. However, Rahimi and Recht (2008) obtained finite-dimensional feature maps approximating the kernels \(k_{\text {RBF}}\) and \(k_{\varDelta }\) of Eq. (9). Similar results are known for other popular kernels for vectorial data like the Jaccard (Vedaldi and Zisserman 2012) and the Laplacian kernel (Andoni 2009).
In the following we approximate \(k_V(v,w)\) in Eq. (6) by \(\langle {\widetilde{\phi }}^\text {V}(v), {\widetilde{\phi }}^\text {V}(w) \rangle \), where \({\widetilde{\phi }}^\text {V}\) is finite-dimensional, approximative mapping, such that with probability \((1-\delta )\) for \(\delta \in (0,1)\)
for any \(\varepsilon > 0\), and derive a finite-dimensional, approximative feature map for weighted vertex kernels.
Proposition 4
Let \(K^\text {WV}\) be a weighted vertex kernel and let \(\mathcal {D}\) be a non–empty finite set of graphs. Further, let \(\phi ^W\) be a feature map for \(k_W\) and let \({\widetilde{\phi }}^{\text {V}}\) be an approximative mapping for \(k_V\) according to Eq. (11). Then we can compute an approximative feature map \({\widetilde{\phi }}^\text {WV}\) for \(K^\text {WV}\) such that with any constant probability
for any \(\lambda > 0\).
Proof
By inequality (11) we get that for every pair of vertices in the data set \(\mathcal {D}\) with any constant probability
By the above, the accumulated error is
Hence, the result follows by setting \(\varepsilon = \lambda /(k^{\text {W}}_{\max } \cdot |V_{\max }|^2)\), where \(k^{\text {W}}_{\max }\) is the maximum value attained by the kernel \(k^{\text {W}}\) and \(|V_{\max }|\) is the maximum number of vertices over the whole data set. \(\square \)
5.2 Fixed length walk kernels
In contrast to the classical walk based graph kernels, fixed length walk kernels take only walks up to a certain length into account. Such kernels have been successfully used in practice (Borgwardt et al. 2005; Harchaoui and Bach 2007) and are not susceptible to the phenomenon of halting (Sugiyama and Borgwardt 2015). We propose an explicit and implicit computation scheme for fixed length walk kernels supporting arbitrary vertex and edge kernels. Our implicit computation scheme is based on product graphs and benefits from sparse vertex and edge kernels. Previously no algorithms based on explicit mapping for computation of walk-based kernels have been proposed. For graphs with discrete labels, we identify the label diversity and walk lengths as key parameters affecting the running time. This is confirmed experimentally in Sect. 6.
5.2.1 Basic definitions
A fixed length walk kernel measures the similarity between graphs based on the similarity between all pairs of walks of length \(\ell \) contained in the two graphs. A walk of length \(\ell \) in a graph G is a sequence of vertices and edges \((v_0,e_1,v_1,\dots ,e_\ell ,v_\ell )\) such that \(e_i= v_{i-1}v_i \in E(G)\) for \(i \in \{1, \dots , \ell \}\). We denote the set of walks of length \(\ell \) in a graph G by \(\mathcal {W}_\ell (G)\).
Definition 2
(\(\ell \)-walk kernel) The \(\ell \)-walk kernel between two attributed graphs G and H in \({{\mathcal {G}}} \) is defined as
where \(k_W\) is a kernel between walks.
Definition 2 is very general and does not specify how to compare walks. An obvious choice is to decompose walks and define \(k_W\) in terms of vertex and edge kernel functions, denoted by \(k_V\) and \(k_E\), respectively. We consider
where \(w = (v_0,e_1,\dots ,v_\ell )\) and \(w' = (v'_0,e'_1,\dots ,v'_\ell )\) are two walks.Footnote 2 Assume the graphs in a data set have simple vertex and edge labels \(\tau :V \uplus E \rightarrow \mathcal {L}\). An appropriate choice then is to use the Dirac kernel for both, vertex and edge kernels, between the associated labels. In this case two walks are considered equal if and only if the labels of all corresponding vertices and edges are equal. We refer to this kernel by
where \(k_\delta \) is the Dirac kernel. For graphs with continuous or multi-dimensional annotations this choice is not appropriate and \(k_V\) and \(k_E\) should be selected depending on the application-specific vertex and edge attributes.
A variant of the \(\ell \)-walk kernel can be obtained by considering all walks up to length \(\ell \).
Definition 3
(Max-\(\ell \)-walk kernel) The Max-\(\ell \)-walk kernel between two attributed graphs G and H in \({{\mathcal {G}}} \) is defined as
where \(\lambda _0, \dots , \lambda _\ell \in \mathbb {R}_{\ge 0} \) are weights.
This kernel is referred to as k-step random walk kernel by Sugiyama and Borgwardt (2015). In the following we primary focus on the \(\ell \)-walk kernel, although our algorithms and results can be easily transferred to the Max-\(\ell \)-walk kernel.
5.2.2 Walk and convolution kernels
We show that the \(\ell \)-walk kernel is p.s.d. if \(k_W\) is a valid kernel by seeing it as an instance of an R-convolution kernel. We use this fact to develop an algorithm for explicit mapping based on the ideas presented in Sect. 4.4.
Proposition 5
The \(\ell \)-walk kernel is positive semidefinite if \(k_W\) is defined according to Eq. (14) and \(k_V\) and \(k_E\) are valid kernels.
Proof
The result follows from the fact that the \(\ell \)-walk kernel can be seen as an instance of an R-convolution kernel, cf. Definition 1, where graphs are decomposed into walks. Let \(w = (v_0, e_1, v_1, \dots , e_\ell , v_\ell ) = (x_0, \dots , x_{2\ell })\) and \(w' = (x'_0, \dots , x'_{2\ell })\) be two walks and \(\kappa (w, w')=\prod _{i=0}^{2\ell } \kappa _i(x_i,x'_i)\) with
for \(i \in \{0,\dots ,2\ell \}\), then \(k_W = \kappa \). This implies that the \(\ell \)-walk kernel is a valid kernel if \(k_V\) and \(k_E\) are valid kernels. \(\square \)
Since kernels are closed under taking linear combinations with non-negative coefficients, see Sect. 4, we obtain the following corollary.
Corollary 4
The Max-\(\ell \)-walk kernel is positive semidefinite.
5.2.3 Implicit kernel computation
An essential part of the implicit computation scheme is the generation of the product graph that is then used to compute the \(\ell \)-walk kernel.
Computing direct product graphs In order to support graphs with arbitrary attributes, vertex and edge kernels \(k_V\) and \(k_E\) are considered as part of the input. Product graphs can be used to represent these kernel values between pairs of vertices and edges of the input graphs in a compact manner. We avoid to create vertices and edges that would represent incompatible pairs with kernel value zero. The following definition can be considered a weighted version of the direct product graph introduced by Gärtner et al. (2003) for kernel computation.Footnote 3
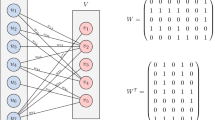
Definition 4
(Weighted direct product graph) For two attributed graphs \(G=(V,E)\), \(H=(V',E')\) and given vertex and edge kernels \(k_V\) and \(k_E\), the weighted direct product graph (WDPG) is denoted by \(G \times _{w}H = (\mathcal {V}, \mathcal {E},w)\) and defined as
Here \([\mathcal {V}]^2\) denotes the set of all 2-element subsets of \(\mathcal {V}\).
Two attributed graphs G (a) and H (b) and their weighted direct product graph \(G \times _{w}H\) (c). We assume the vertex kernel to be the Dirac kernel and \(k_E\) to be 1 if edge labels are equal and \(\frac{1}{2}\) if one edge label is “=” and the other is “-”. Thin edges in \(G \times _{w}H\) represent edges with weight \(\frac{1}{2}\), while all other edges and vertices have weight 1
An example with two graphs and their weighted direct product graph obtained for specific vertex and edge kernels is shown in Fig. 1. Algorithm 1 computes a weighted direct product graph and does not consider edges between pairs of vertices \((v,v')\) that have been identified as incompatible, i.e., \(k_V(v,v')=0\).

Since the weighted direct product graph is undirected, we must avoid that the same pair of edges is processed twice. Therefore, we suppose that there is an arbitrary total order \(\prec \) on the vertices \(\mathcal {V}\), such that for every pair \((u,s),(v,t)\in \mathcal {V}\) either \((u,s) \prec (v,t)\) or \((v,t) \prec (u,s)\) holds. In line 8 we restrict the edge pairs that are compared to one of these cases.
Proposition 6
Let \(n=|V(G)|\), \(n'=|V(H)|\) and \(m=|E(G)|\), \(m'=|E(H)|\). Algorithm 1 computes the weighted direct product graph in time \(\mathcal {O}(n n' {\mathsf {T}}_{V} + m m' {\mathsf {T}}_{E})\), where \({\mathsf {T}}_{V}\) and \({\mathsf {T}}_{E}\) is the running time to compute vertex and edge kernels, respectively.
Note that in case of a sparse vertex kernel, which yields zero for most of the vertex pairs of the input graph, \(|V(G \times _{w}H)| \ll |V(G)| \cdot |V(H)|\) holds. Algorithm 1 compares two edges by \(k_E\) only in case of matching endpoints (cf. lines 7, 8), therefore in practice the running time to compare edges (line 7–13) might be considerably less than suggested by Proposition 6. We show this empirically in Sect. 6.4. In case of sparse graphs, i.e., \(|E|=\mathcal {O}(|V|)\), and vertex and edge kernels which can be computed in time \(\mathcal {O}(1)\) the running time of Algorithm 1 is \(\mathcal {O}(n^2)\), where \(n=\max \{|V(G)|,|V(H)|\}\).
Counting weighted walks Given an undirected graph G with adjacency matrix \(\mathbf {A}\), let \(a^\ell _{ij}\) denote the element at (i, j) of the matrix \(\mathbf {A}^\ell \). It is well-known that \(a^\ell _{ij}\) is the number of walks from vertex i to j of length \(\ell \). The number of \(\ell \)-walks of G consequently is \(\sum _{i,j} a^\ell _{i,j} = {\mathbf {1}}^\top \mathbf {A}^\ell {\mathbf {1}} = {\mathbf {1}}^\top \mathbf {r}_\ell \), where \(\mathbf {r}_\ell = \mathbf {A} \mathbf {r}_{\ell -1}\) with \(\mathbf {r}_0 = {\mathbf {1}}\). The ith element of the recursively defined vector \(\mathbf {r}_\ell \) is the number of walks of length \(\ell \) starting at vertex i. Hence, we can compute the number of \(\ell \)-walks by computing either matrix powers or matrix-vector products. Note that even for sparse (connected) graphs \(\mathbf {A}^\ell \) quickly becomes dense with increasing walk length \(\ell \). The \(\ell \)th power of an \(n \times n\) matrix \(\mathbf {A}\) can be computed naïvely in time \(\mathcal {O}(n^\omega \ell )\) and \(\mathcal {O}(n^\omega \log \ell )\) using exponentiation by squaring, where \(\omega \) is the exponent of matrix multiplication. The vector \(\mathbf {r}_\ell \) can be computed by means of matrix-vector multiplications, where the matrix \(\mathbf {A}\) remains unchanged over all iterations. Since direct product graphs tend to be sparse in practice, we propose a method to compute the \(\ell \)-walk kernel that is inspired by matrix-vector multiplication.
In order to compute the \(\ell \)-walk kernel we do not want to count the walks, but sum up the weights of each walk, which in turn are the product of vertex and edge weights. Let \(k_W\) be defined according to Eq. (14), then we can formulate the \(\ell \)-walk kernel as
where \(r_\ell \) is determined recursively according to
Note that \(r_i\) can as well be formulated as matrix-vector product. We present a graph-based approach for computation akin to sparse matrix-vector multiplication, see Algorithm 2.

Theorem 1
Let \(n=|\mathcal {V}|\), \(m=|\mathcal {E}|\). Algorithm 2 computes the \(\ell \)-walk kernel in time \(\mathcal {O}(n+\ell (n+m) + {\mathsf {T}}_{\mathrm{WDPG}})\), where \({\mathsf {T}}_{\mathrm{WDPG}}\) is the time to compute the weighted direct product graph.
Note that the running time depends on the size of the product graph and \(n \ll |V(G)| \cdot |V(H)|\) and \(m \ll |E(G)| \cdot |E(H)|\) is possible as discussed in Sect. 5.2.3.
The Max-\(\ell \)-walk kernel is the sum of the j-walk kernels with \(j \le \ell \) and, hence, with Eq. (17) we can also formulate it recursively as
This value can be obtained from Algorithm 2 by simply changing the return statement in line 9 according to the right-hand side of the equation without affecting the asymptotic running time.
5.2.4 Explicit kernel computation
We have shown in Sect. 5.2.2 that \(\ell \)-walk kernels are R-convolution kernels. Therefore, we can derive explicit feature maps with the techniques introduced in Sect. 4. Provided that we know explicit feature maps for the vertex and edge kernel, we can derive explicit feature maps for the kernel on walks and obtain an explicit computation scheme by enumerating all walks. We propose a more elaborated approach that avoids enumeration and exploits the simple composition of walks.
To this end, we consider Eqs. (17), (18) and (19) of the recursive product graph based implicit computation. Let us assume that \(\phi ^V\) and \(\phi ^E\) are feature maps of the kernels \(k_V\) and \(k_E\), respectively. Then we can derive a feature map \(\varPhi _i\), such that \(\langle \varPhi _i(u), \varPhi _i(u') \rangle = r_i(u,u')\) for all \((u,u') \in V(G) \times V(H)\)Footnote 4 as follows using the results of Sect. 4.
From these, the feature map \(\phi ^=_\ell \) of the \(\ell \)-walk kernel is obtained according to
Algorithm 3 provides the pseudo code of this computation. We can easily derive a feature map of the Max-\(\ell \)-walk kernel from the feature maps of all i-walk kernels with \(i \le \ell \), cf. Proposition 2.

The dimension of the feature space and the density of feature vectors depends multiplicative on the same properties of the feature vectors of \(k_V\) and \(k_E\). For non-trivial vertex and edge kernels explicit computation of the \(\ell \)-walk kernel is likely to be infeasible in practice. Therefore, we now consider graphs with simple labels from the alphabet \(\mathcal {L}\) and the kernel \(k^\delta _W\) given by Eq. (15). Following Gärtner et al. (2003) we can construct a feature map in this case, where the the features are sequences of labels associated with walks. As we will see later, this feature space is indeed obtained with Algorithm 3. A walk w of length \(\ell \) is associated with a label sequence \(\tau (w)=(\tau (v_0),\tau (e_1),\dots ,\tau (v_\ell )) \in \mathcal {L}^{2\ell +1}\). Moreover, graphs are decomposed into walks and two walks w and \(w'\) are considered equivalent if and only if \(\tau (w) = \tau (w')\). This gives rise to the feature map \(\phi ^=_\ell \), where each component is associated with a label sequence \(s \in \mathcal {L}^{2\ell +1}\) and counts the number of walks \(w \in \mathcal {W}_\ell (G)\) with \(\tau (w) = s\). Note that the obtained feature vectors have \(|\mathcal {L}|^{2\ell +1}\) components, but are typically sparse. In fact, Algorithm 3 constructs this feature map. We assume that \(\phi ^V(v)\) and \(\phi ^E(uv)\) have exactly one non-zero component associated with the label \(\tau (v)\) and \(\tau (uv)\), respectively. Then the single non-zero component of \(\phi ^V(u) \otimes \phi ^E(uv) \otimes \phi ^V(v)\) is associated with the label sequence \(\tau (w)\) of the walk \(w=(u, uv, v)\). A walk of length \(\ell \) can be decomposed into a walk of length \(\ell -1\) with an additional edge and vertex added at the front. This allows to obtain the number of walks of length \(\ell \) with a given label sequence starting at a fixed vertex v by concatenating \((\tau (v),\tau (vu))\) with all label sequences for walks starting from a neighbor u of the vertex v. This construction is applied in every iteration of the outer for-loop in Algorithm 3 and the feature vectors \(\varPhi _i\) are easy to interpret. Each component of \(\varPhi _i(v)\) is associated with a label sequence \(s \in \mathcal {L}^{2i+1}\) and counts the walks w of length i starting at v with \(\tau (w)=s\).
We consider the running time for the case, where graphs have discrete labels and the kernel \(k^\delta _W\) is given by Eq. (15).
Theorem 2
Given a graph G with \(n=|V(G)|\) vertices and \(m=|E(G)|\) edges, Algorithm 3 computes the \(\ell \)-walk kernel feature vector \(\phi ^=_\ell (G)\) in time \(\mathcal {O}(n+\ell (n+m)s)\), where s is the maximum number of different label sequences of \((\ell -1)\)-walks staring at a vertex of G.
Assume Algorithm 3 is applied to unlabeled sparse graphs, i.e., \(|E(G)| = \mathcal {O}(|V(G)|)\), then \(s = 1\) and the feature mapping can be performed in time \(\mathcal {O}(n+\ell n)\). This yields a total running time of \(\mathcal {O}(d \ell n + d^2)\) to compute a kernel matrix for d graphs of order n for \(\ell >0\).
5.3 Shortest-path kernel
A classical kernel applicable to attributed graphs is the shortest-path kernel (Borgwardt and Kriegel 2005). This kernel compares all shortest paths in two graphs according to their lengths and the vertex annotation of their endpoints. The kernel was proposed with an implicit computation scheme, but explicit methods of computation have been reported to be used for graphs with discrete labels.
The shortest-path kernel is defined as
where \(k_V\) is a kernel comparing vertex labels of the respective starting and end vertices of the paths. Here, \(d_{uv}\) denotes the length of a shortest path from u to v and \(k_E\) is a kernel comparing path lengths with \(k_E(d_{uv}, d_{wz}) = 0\) if \(d_{uv} = \infty \) or \(d_{wz} = \infty \).
Its computation is performed in two steps (Borgwardt and Kriegel 2005): for each graph G of the data set the complete graph \(G'\) on the vertex set V(G) is generated, where an edge uv is annotated with the length of a shortest path from u to v. The shortest-path kernel then is equivalent to the walk kernel with fixed length \(\ell =1\) between these transformed graphs, where the kernel essentially compares all pairs of edges. The kernel \(k_E\) used to compare path lengths may, for example, be realized by the Brownian Bridge kernel (Borgwardt and Kriegel 2005).
For the application to graphs with discrete labels a more efficient method of computation by explicit mapping has been reported by Shervashidze et al. (2011, Section 3.4.1). When \(k_V\) and \(k_E\) both are Dirac kernels, each component of the feature vector corresponds to a triple consisting of two vertex labels and a path length. This method of computation has been applied in several experimental comparisons, e.g., Kriege and Mutzel (2012) and Morris et al. (2016). This feature map is directly obtained from our results in Sect. 4. It is as well rediscovered from our explicit computation schemes for fixed length walk kernels reported in Sect. 5.2. However, we can also derive explicit feature maps for non-trivial kernels \(k_V\) and \(k_E\). Then the dimension of the feature map increases due to the product of kernels, cf. Eq. 21. We will study this and the effect on running time experimentally in Sect. 6.
5.4 Graphlet, subgraph and subgraph matching kernels
Subgraph or graphlet kernels have been proposed for unlabeled graphs or graphs with discrete labels (Gärtner et al. 2003; Wale et al. 2008; Shervashidze et al. 2009). The subgraph matching kernel has been developed as an extension for attributed graphs (Kriege and Mutzel 2012).
Given two graphs G and H in \(\mathcal {G}\), the subgraph kernel is defined as
where \(k_\simeq :\mathcal {G} \times \mathcal {G} \rightarrow \{0,1\}\) is the isomorphism kernel, i.e., \(k_\simeq (G',H')=1\) if and only if \(G'\) and \(H'\) are isomorphic. A similar kernel was defined by Gärtner et al. (2003) and its computation was shown to be \(\mathsf {NP}\)-hard. However, it is polynomial time computable when considering only subgraphs up to a fixed size. The subgraph kernel, cf. Eq. (22), is easily identified as an instance of the crossproduct kernel, cf. Eq. (2). The base kernel \(k_\simeq \) is not the trivial Dirac kernel, but binary, cf. Sect. 4.1. The equivalence classes induced by \(k_\simeq \) are referred to as isomorphism classes and distinguish subgraphs up to isomorphism. The feature map \(\phi _\subseteq \) of \(k^\subseteq \) maps a graph to a vector, where each component counts the number of occurrences of a specific graph as subgraph in G. Determining the isomorphism class of a graph is known as graph canonization problem and well-studied. By solving the graph canonization problem instead of the graph isomorphism problem we obtain an explicit feature map for the subgraph kernel. Although graph canonization clearly is at least as hard as graph isomorphism, the number of canonizations required is linear in the number of subgraphs, while a quadratic number of isomorphism tests would be required for a single naïve computation of the kernel. The gap in terms of runtime even increases when computing a whole kernel matrix, cf. Sect. 3.
Indeed, the observations above are key to several graph kernels. The graphlet kernel (Shervashidze et al. 2009), also see Sect. 2, is an instance of the subgraph kernel and computed by explicit feature maps. However, only unlabeled graphs of small size are considered by the graphlet kernel, such that the canonizing function can be computed easily. The same approach was taken by Wale et al. (2008) considering larger connected subgraphs of labeled graphs derived from chemical compounds. On the contrary, for attributed graphs with continuous vertex labels, the function \(k_\simeq \) is not sufficient to compare subgraphs adequately. Therefore, subgraph matching kernels were proposed by Kriege and Mutzel (2012), which allow to specify arbitrary kernel functions to compare vertex and edge attributes. Essentially, this kernel considers all mappings between subgraphs and scores each mapping by the product of vertex and edge kernel values of the vertex and edge pairs involved in the mapping. When the specified vertex and edge kernels are Dirac kernels, the subgraph matching kernel is equal to the subgraph kernel up to a factor taking the number of automorphisms between subgraphs into account (Kriege and Mutzel 2012). Based on the above observations explicit mapping of subgraph matching kernels is likely to be more efficient when subgraphs can be adequately compared by a binary kernel.
5.5 Discussion
A crucial observation of our study of feature maps for composed kernels in Sect. 4 is that the number of components of the feature vectors increases multiplicative under taking products of kernels; this also holds in terms of non-zero components. Unless feature vectors have few non-zero components, this operation is likely to be prohibitive in practice. However, if feature vectors have exactly one non-zero component like those associated with binary kernels, taking products of kernels is manageable by sparse data structures.
In this section we have systematically constructed and discussed feature maps of several graph kernels and the observation mentioned above is expected to affect the kernels to varying extents. While weighted vertex kernels do not take products of vertex and edge kernels, the shortest-path kernel and, in particular, the subgraph matching and fixed length walk kernels heavily rely on multiplicative combinations. Considering the relevant special case of a Dirac kernel, which leads to feature vectors with only one non-zero component, the rapid growth due to multiplication is tamed. In this case the number of substructures considered as different according to the vertex and edge kernels determines the number of non-zero components of the feature vectors associated with the graph kernel. The basic characteristics of the considered graph kernels are summarized in Table 2. The sparsity of the feature vectors of the vertex and edge kernels is an important intervening factor, which is difficult to assess theoretically and we proceed by an experimental study.
6 Experimental evaluation
Our goal in this section is to answer the following questions experimentally.
-
Q1
Are approximative explicit feature maps of kernels for attributed graphs competitive in terms of running time and classification accuracy compared to exact implicit computation?
-
Q2
Are exact explicit feature maps competitive for kernels relying on multiplication when the Dirac kernel is used to compare discrete labels? How do the graph properties such as label diversity affect the running time?
-
(a)
How does the fixed length walk kernel behave with regard to these questions and what influence does the walk length have?
-
(b)
Can the same behavior regarding running time be observed for the graphlet and subgraph matching kernel?
-
(a)
6.1 Experimental setup
All algorithms were implemented in Java and the default Java HashMap class was used to store feature vectors. Due to the varied memory requirements of the individual series of experiments, different hardware platforms were used in Sects. 6.2, 6.4 and 6.5. In order to compare the running time of both computational strategies systematically without the dependence on one specific kernel method, we report the running time to compute the quadratic kernel matrices, unless stated otherwise. We performed classification experiments using the C-SVM implementation LIBSVM (Chang and Lin 2011). We report mean prediction accuracies obtained by 10-fold cross-validation repeated 10 times with random fold assignments. Within each fold all necessary parameters were selected by cross-validation based on the training set. This includes the regularization parameter C selected from \(\{10^{-3}, 10^{-2}, \dots , 10^3\}\), all kernel parameters, where applicable, and whether to normalize the kernel matrix.
6.1.1 Data sets
We performed experiments on synthetic and real-world data sets from different domains, see Table 3 for an overview on their characteristics. All data sets can be obtained from our publicly available collection (Kersting et al. 2016) unless the source is explicitly stated.
Small molecules Molecules can naturally be represented by graphs, where vertices represent atoms and edges represent chemical bonds. Mutag is a data set of chemical compounds divided into two classes according to their mutagenic effect on a bacterium. This small data set is commonly used in the graph kernel literature. In addition we considered the larger data set U251, which stems from the NCI Open Database provided by the National Cancer Institute (NCI). In this data set the class labels indicate the ability of a compound to inhibit the growth of the tumor cell line U251. We used the data set processed by Swamidass et al. (2005), which is publicly available from the ChemDB website.Footnote 5
MacromoleculesEnzymes and Proteins both represent macromolecular structures and were obtained from Borgwardt et al. (2005) and Feragen et al. (2013). The following graph model has been employed. Vertices represent secondary structure elements (SSE) and are annotated by their type, i.e., helix, sheet or turn, and a rich set of physical and chemical attributes. Two vertices are connected by an edge if they are neighbors along the amino acid sequence or one of three nearest neighbors in space. Edges are annotated with their type, i.e., structural or sequential. In Enzymes each graph is annotated by an EC top level class, which reflects the chemical reaction the enzyme catalyzes, Proteins is divided into enzymes and non-enzymes.
Synthetic graphs The data sets SyntheticNew and Synthie were synthetically generated to obtain classification benchmarks for graph kernels with attributes. We refer the reader to the publications Feragen et al. (2013)Footnote 6 and Morris et al. (2016), respectively, for the details of the generation process. Additionally, we generated new synthetic graphs in order to systematically vary graph properties of interest like the label diversity, which we expect to have an effect on the running time according to our theoretical analysis.
6.2 Approximative explicit feature maps of kernels for attributed graphs (Q1)
We have derived explicit computation schemes of kernels for attributed graphs, which have been proposed with an implicit method of computation. Approximative explicit computation is possible under the assumption that the kernel for the attributes can be approximated by explicit feature maps. We compare both methods of computation w.r.t. their running time and the obtained classification accuracy on the four attributed graph data sets Enzymes, Proteins, SyntheticNew and Synthie. Since the discrete labels alone are often highly informative, we ignored discrete labels if present and considered the real-valued vertex annotations only in order to obtain challenging classification problems. All attributes where dimension-wise linearly scaled to the range [0, 1] in a preprocessing step.
6.2.1 Method
We employed three kernels for attributed graphs: the shortest-path kernel, cf. Sect. 5.3, the GraphHopper and GraphInvariant kernel as described in Sect. 5.1.1. Preliminary experiments with the subgraph matching kernel showed that it cannot be computed by explicit feature maps for non-trivial subgraph sizes due to its high memory consumption. The same holds for fixed length walk kernels with walk length \(\ell \ge 3\). Therefore, we did not consider these kernels any further regarding Q1, but investigate them for graphs with discrete labels in Sects. 6.4 and 6.5 to answer Q2a and Q2b.
For the shortest-path kernel we used the Dirac kernel to compare path lengths and selected the number of Weisfeiler-Lehman refinement steps for the GraphInvariant kernel from \(h \in \{0, \dots , 7\}\). For the comparison of attributes we employed the dimension-wise product of the hat kernel \(k_\varDelta \) as defined in Eq. (9) choosing \(\delta \) from \(\{0.2, 0.4, 0.6, 0.8, 1.0, 1.5, 2.0\}\). The three kernels were computed functionally employing this kernel as a base line. We obtained approximate explicit feature maps for the attribute kernel by the method of Rahimi and Recht (2008) and used these to derive approximate explicit feature maps for the graph kernels. We varied the number of random binning features from \(\{1, 2, 4, 8, 16, 32, 64\}\), which corresponds to the number of non-zero components of the feature vectors for the attribute kernel and controls its approximation quality. Please note that the running time is effected by the kernel parameters, i.e., \(\delta \) of Eq. (9) and the number h of Weisfeiler-Lehman refinement steps for GraphInvariant. Therefore, in the following we report the running times for fixed values \(\delta =1\) and \(h=3\), which were selected frequently by cross-validation.
All experiments were conducted using Oracle Java v1.8.0 on an Intel Xeon E5-2640 CPU at 2.5 GHz with 64 GB of RAM using a single processor only.
Running times and classification accuracies of graph kernels approximated by explicit feature maps with \(2^i\), \(i \in \{0,\dots ,4\}\), iterations of random binning. The results of exact implicit computation are shown as a base line (left y-axes shows the running time in seconds, right y-axes the accuracy in percent; GH—GraphHopper, GI—GraphInvariant, SP—ShortestPath)
6.2.2 Results and discussion
We were not able to compute the shortest-path kernel by explicit feature maps with more than 16 iterations of binning for the base kernel on Enzymes and Proteins and no more than 4 iterations on Synthie and SyntheticNew with 64 GB of main memory. The high memory consumption of this kernel is in accordance with our theoretical analysis, since the multiplication of vertex and edge kernels drastically increases the number of non-zero components of the feature vectors. This problem does not effect the two weighted vertex kernels to the same extent. We observed the general trend that the memory consumption and running time increases with small values of \(\delta \). This is explained by the fact that the number of components of the feature vectors of the vertex kernels increases in this case. Although the number of non-zero components does not increase for these feature vectors, it does for the graph kernel feature vectors, since the number of vertices with attributes falling into different bins increases.
The results on running time and accuracy are summarized in Fig. 2. For the two data sets Enzymes and Synthie we observe that the classification accuracy obtained by the approximate explicit feature maps approaches the accuracy obtained by the exact method with increasing number of binning iterations. For the other two data sets the accuracy does not improve with the number of iterations. For Proteins the kernels obtained with a single iteration of binning, i.e., essentially applying a Dirac kernel, achieve an accuracy at the same level as the exact kernel obtained by implicit computation. This suggests that for this data set a trivial comparison of attributes is sufficient or that the attributes are not essential for classification at all. For SyntheticNew the kernels using a single iteration of binning are even better than the exact kernel, but get worse as the number of iterations increases. One possible explanation for this is that the vertex kernel used is not a good choice for this data set.
With few iteration of binning the explicit computation scheme is always faster than the implicit computation. The growth in running time with increasing number of binning iterations for the vertex kernel varies between the graph kernels. Approximating the GraphHopper kernel by explicit feature maps with 64 binning iteration for the vertex kernel leads to a running time similar to the one required for its exact implicit computation on all data sets with exception of SyntheticNew. On this data set explicit computation remains faster. For GraphInvariant explicit feature maps lead to a running time which is orders of magnitude lower than implicit computation. Although both, GraphHopper and GraphInvariant are weighted vertex kernels, this difference can be explained by the number of non-zero components in the feature vectors of the weight kernel. We observe that GraphInvariant clearly provides the best classification accuracy for two of the four data sets and is competitive for the other two. At the same time GraphInvariant can be approximated very efficiently by explicit feature maps. Therefore, even for attributed graphs effective and efficient graph kernels can be obtained from explicit feature maps by our approach.
6.3 Kernels for graphs with discrete labels (Q2)
In order to get a first impression of the runtime behavior of explicit and implicit computation schemes on graphs with discrete labels, we computed the kernel matrix for the standard data sets ignoring the attributes, if present. The experiments were conducted using Java OpenJDK v1.7.0 on an Intel Core i7-3770 CPU at 3.4 GHz (Turbo Boost disabled) with 16 GB of RAM using a single processor only. The reported running times are average values over 5 runs.
The results are summarized in Table 4. For the shortest-path kernel explicit mapping clearly outperforms implicit computation by several orders of magnitude with respect to running time. This is in accordance with our theoretical analysis and our results suggest to always use explicit computation schemes for this kernel whenever a Dirac kernel is adequate for label and path length comparison. In this case memory consumption is unproblematic, in contrast to the setting discussed in Sect. 6.2.
Note that the explicit computation of the fixed length random walk kernel is extremely efficient on SyntheticNew and Synthie, which have uniform discrete labels, but strongly depends on the walk length for the other data sets. This observation is investigated in detail in Sect. 6.4. The running times of the connected subgraph matching kernel and the graphlet kernel are studied exhaustively in Sect. 6.5.
6.4 Fixed length walk kernels for graphs with discrete labels (Q2a)
Our comparison in Sect. 6.2 showed that computation by explicit feature maps becomes prohibitive when vertex and edge kernels with feature vectors having multiple non-zero components are multiplied. This is even observed for the shortest-path kernel, which applies a walk kernel of fixed length one. Therefore, we study the implicit and explicit computation schemes of the fixed length walk kernel on graphs with discrete labels, which are compared by the Dirac kernel, cf. Eq. (15). Since both computation schemes produce the same kernel matrices, our main focus in this section is on running times.
The discussion of running times for walk kernels in Sects. 5.2.3 and 5.2.4 suggested that
-
(i)
implicit computation benefits from sparse vertex and edge kernels,
-
(ii)
explicit computation is promising for graphs with a uniform label structure, which exhibit few different features, and then scales to large data sets.
We experimentally analyze this trade-off between label diversity and running time for synthetic and real-world data sets ignoring any attributes, if present. Finally, we use our walk kernels to compare graphs after applying different levels of label refinement using the Weisfeiler-Lehman method. We used the same experimental method as reported in Sect. 6.3.
6.4.1 Synthetic data sets
In order to systematically vary the label diversity we generated synthetic graphs by the following procedure: the number of vertices was determined by a Poisson distribution with mean 20. Edges were inserted between a pair of vertices with probability 0.1. The label diversity depends on the parameter \(p_V\). Edges were uniformly labeled; a vertex obtained the label 0 with probability \(1-p_V\). Otherwise the labels 1 or 2 were assigned with equal probability. In addition, we vary the data set size d between 100 and 300 adding 20 randomly generated graphs in each step.
The results are depicted in Fig. 3, where a label diversity of 50 means that \(p_V=0.5\). Figure 3a shows that the running time for implicit computation increases with the data set size and decreases with the label diversity. This observation is in accordance with our hypotheses. When the label diversity increases, there are less compatible pairs of vertices and the weighted direct product graph becomes smaller. Consequently, its computation and the counting of weighted walks require less running time. For explicit computation we observe a different trend: while the running time increases with the size of the data set, the approach is extremely efficient for graphs with uniform labels (\(p_V=0\)) and becomes slower when the label diversity increases, cf. Fig. 3b. Combining both results, cf. Fig. 3c, shows that both approaches yield the same running time for a label diversity of \(p_V \approx 0.3\), while for higher values of \(p_V\) implicit computation is preferable and explicit otherwise.
6.4.2 Molecular data sets
In the previous section we have observed how both approaches behave when the label diversity is varied. We use the data set U251 of graphs derived from small molecules to analyze the running time on a real-world data set with a predetermined label diversity. Vertex labels correspond to the atom types and edge labels represent single, double, triple and aromatic bonds, respectively. This time we vary the walk length and the data set size by starting with a random subset and adding additional graphs that were selected randomly from the remaining graphs of the data set.
Figure 4a shows that the running time of the implicit computation scheme heavily depends on the size of the data set. The increase with the walk length is less considerable. This can be explained by the time \({\mathsf {T}}_{\mathrm{WDPG}}\) required to compute the product graph, which is always needed independent of the walk length. For short walks explicit computation is very efficient, even for larger data sets, cf. Fig. 4b. However, when a certain walk length is reached the running time increases drastically. This can be explained by the growing number of different label sequences. Notably for walks of length 8 and 9 the running time also largely increases with the data set size. This indicates that the time \({\mathsf {T}}_{\text {dot}}\) has a considerable influence on the running time. In the following section we analyze the running time of the different procedures for the two algorithms in more detail. Figure 4c shows that for walk length up to 7 explicit computation beats implicit computation on the molecular data set.
6.4.3 Enzymes and Mutag
We have shown that up to a certain walk length explicit computation is more efficient than implicit computation. We want to clarify the relation between the walk length and the prediction accuracy in a classification task. In addition, we analyze the ratio between the time \({\mathsf {T}}_\phi \) for computing the explicit mapping and \({\mathsf {T}}_{\text {dot}}\) for taking dot products. For the implicit computation scheme we want to clarify the running time of \({\mathsf {T}}_{\mathrm{WDPG}}\) and the time required for counting weighted walks. We apply both algorithms to two widely used data sets, Mutag and Enzymes, and vary the walk length, see Table 3 for details on these data sets.
Figure 5 shows the running time of both algorithms depending on the walk length and gives the time for product graph computation and explicit mapping, respectively. In addition, the prediction accuracy is presented. For both data sets we observe that up to a walk length of 7 explicit mapping is more efficient. Notably a peak of the accuracy is reached for walk length smaller than 7 in both cases. For the Mutag data set walks of length 3 provide the best results and walks of length 6 for the Enzymes data set, i.e., in both cases explicit mapping should be preferred when computing a walk kernel of fixed length. The running time of the product graph computation is constant and does not depend on the walk length. For explicit mapping the time required to compute the dot product becomes dominating when the walk length is increased. This can be explained by the fact that the generation of the kernel matrix involves a quadratic number of dot product computations. Note that the given times include a quadratic number of product graph computations while the times for generating the feature vectors include only a linear number of operations.
As a side note, we also compared the accuracy of the fixed length walk kernels to the accuracy reached by the geometric random walk kernel (GRW) according to Gärtner et al. (2003), which considers arbitrary walk lengths. The parameter \(\gamma \) of the geometric random walk kernel was selected by cross-validation from \(\{10^{-5}, 10^{-4},\dots ,10^{-2}\}\). We observed that the accuracy of the fixed length walk kernel is competitive on the Mutag data set (GRW 87.3), and considerably better on the Enzymes data set (GRW 31.6), cf. Fig. 5. This is remarkable, since the fixed length walk kernel yields best results with walk length 6, for which it is efficiently computed by explicit mapping. However, this is not possible for the geometric random walk kernel. For a more detailed discussion and comparison between fixed length walk kernels and the geometric random walk kernel we refer the reader to Sugiyama and Borgwardt (2015), which appeared after the conference publication (Kriege et al. 2014).
6.4.4 Weisfeiler-Lehman label refinement
Walk kernels have been successfully combined with label refinement techniques (Mahé et al. 2004). We employ the Weisfeiler-Lehman label refinement (WL) as described in Sect. 5.1.1. To further analyze the sensitivity w.r.t. label diversity, we again use the Enzymes data set, which consists of graphs with three vertex and two edge labels initially. We apply our algorithms for explicit and implicit computation after 0 to 3 iterations of WL, see Fig. 6.
If no refinement is applied, the explicit mapping approach beats the product graph based algorithm for the used walk lengths. However, as soon as a single iteration of label refinement is performed, the product graph based algorithm becomes competitive for walk length 0 and 1 and outperforms the explicit mapping approach for higher walk lengths. The running times do not change substantially for more iterations of refinement. This indicates that a single iteration of Weisfeiler-Lehman refinement results in a high label diversity that does not increase considerably for more iterations on the Enzymes data set. When using our walk-based kernel as base kernel of a Weisfeiler-Lehman graph kernel (Shervashidze et al. 2011), our observation suggests to start with explicit computation and switch to the implicit computation scheme after few iterations of refinement.
6.5 Subgraph kernels for graphs with discrete labels (Q2b)
In this section we experimentally compare the running time of the subgraph matching and the subgraph (or graphlet) kernel as discussed in Sect. 5.4. The explicit computation scheme, which is possible for graphs with discrete labels compared by the Dirac kernel, is expected to be favorable.
6.5.1 Method
We have reimplemented a variation of the graphlet kernel taking connected induced subgraphs with three vertices and discrete vertex and edge labels into account. The only possible features are triangles and paths of length two. Graph canonization is realized by selecting the lexicographically smallest string obtained by traversing the graph and concatenating the observed labels. Our implementation is similar to the approach used by Shervashidze et al. (2011) as extension of the original graphlet kernel (Shervashidze et al. 2009) to the domain of labeled graphs. We refer to this method as graphlet kernel in the following. We compared the graphlet kernel to the connected subgraph matching kernel taking only connected subgraphs on three vertices into account. In order not to penalize the running time of the connected subgraph matching kernel by additional automorphism computations, the weight function does not consider the number of automorphisms (Kriege and Mutzel 2012, Theorem 2) and, consequently, not the same kernel values are computed.
The experiments were conducted using Sun Java JDK v1.6.0 on an Intel Xeon E5430 machine at 2.66 GHz with 8 GB of RAM using a single processor only. The reported running times are average values over 5 runs.
6.5.2 Results and discussion
For real-world instances we observed that explicit computation outperforms implicit computation by several orders of magnitude, cf. Table 4. This in accordance with our theoretical analysis. However, the practical considerations suggest that explicit and implicit computation behave complementary and subgraph matching kernels become competitive if a sufficient small and sparse weighted product graph is generated, which occurs for graphs with increasing label diversity as for the walk-based kernels. Hence, we randomly generated graphs with the following procedure: the number of vertices was determined by a Poisson distribution with mean 60. Edges were inserted between a pair of vertices with probability 0.5. Labels for vertices and edges were assigned with equal probability, whereas the size of the label alphabet \(\mathcal {L}=\mathcal {L}_V=\mathcal {L}_E\) is varied from 1, i.e., uniform labels, to 65. Note that the graphs obtained by this procedure have different characteristics than those used to show the computational phase transition for walk-based kernels. We vary the data set size d between 100 and 300 adding 50 randomly generated graphs in each step and analyze the running time to compute the \(d \times d\) kernel matrix. For the subgraph matching kernel we used the Dirac kernel as vertex and edge kernel.
Figure 7 shows a computational phase transition: for the synthetic data set the subgraph matching kernel is more efficient than the graphlet kernel for instances with 20–30 different labels and its running time increases drastically when the number of labels decreases. The graphlet kernel in contrast is more efficient for graphs with uniform or few labels. For more than 10 different labels, there is only a moderate increase in running time. This can be explained by the fact that the number of features contained in the graphs does not increase considerably as soon as a certain number of different labels is reached. The enumeration of triangles dominates the running time for this relatively dense synthetic data set. The running time behavior of the subgraph matching kernel is as expected and is directly related to the size and number of edges in the weighted association graph.
Our synthetic data set differs from typical real-world instances, since we generated dense graphs with many different labels, which are assigned uniformly at random. For real-world data sets the graphlet kernel consistently outperforms the subgraph matching kernel by orders of magnitude. It would be interesting to further investigate where this computational phase transition occurs for larger subgraphs and to analyze if the implicit computation scheme then becomes competitive for instances of practical relevance. This requires the implementation of non-trivial graph canonization algorithms and remains future work. The results we obtained clearly suggest to prefer the explicit computation schemes when no flexible scoring by vertex and edge kernels is required.
7 Conclusion
The breadth of problems requiring to deal with graph data is growing rapidly and graph kernels have become an efficient and widely used method for measuring similarity between graphs. Highly scalable graph kernels have recently been proposed for graphs with thousands and millions of vertices based on explicit graph feature maps. Implicit computation schemes are used for kernels with a large number of possible features such as walks and when graphs are annotated by continuous attributes.
To set the stage for the experimental comparison, we actually made several contributions to the theory and algorithmics of graph kernels. We presented a unified view on implicit and explicit graph features. More precisely, we derived explicit feature maps from the implicit feature space of convolution kernels and analyzed the circumstances rendering this approach feasible in practice. Using these results, we developed explicit computation schemes for random walk kernels (Gärtner et al. 2003; Vishwanathan et al. 2010), subgraph matching kernels (Kriege and Mutzel 2012), and shortest-path kernels (Borgwardt and Kriegel 2005). Moreover, we introduced weighted vertex kernels and derived explicit feature maps. As a result of this we obtained approximate feature maps for state-of-the-art kernels for graphs with continuous attributes such as the GraphHopper kernel (Feragen et al. 2013). For fixed length walk kernels we have developed implicit and explicit computation schemes and analyzed their running time. Our theoretical results have been confirmed experimentally by observing a computational phase transition with respect to label diversity and walk lengths.
We have shown that kernels composed by multiplication of non-trivial base kernels may lead to a rapid growth of the number of non-zero components in the feature vectors, which renders explicit computation infeasible. One approach to alleviate this in future work is to introduce sampling or hashing to obtain compact feature representations in such cases, e.g., following the work by Shi et al. (2009).
Notes
Note that we may consider every kernel \(\kappa _i\) on \(\mathcal {R}_i\) as kernel \(\kappa '_i\) on \(\mathcal {R}\) by defining \(\kappa '_i(x,y)=\kappa _i(x_i,y_i)\).
The same idea to compare walks was proposed by Kashima et al. (2003) as part of the marginalized kernel between labeled graphs.
Note that we consider undirected graphs while Gärtner et al. (2003) refers to directed graphs.
We assume \(r_i(u,u')=0\) for \((u,u') \notin V(G \times _{w}H)\) .
We used the updated version of the data set Synthetic published together with the Erratum to Feragen et al. (2013).
References
Andoni A (2009) Nearest neighbor search: the old, the new, and the impossible. Ph.D. thesis, MIT
Bai L, Rossi L, Zhang Z, Hancock ER (2015) An aligned subtree kernel for weighted graphs. In: Proceedings of the thirty-second international conference on machine learning, pp 30–39
Borgwardt KM, Kriegel HP (2005) Shortest-path kernels on graphs. In: Proceedings of the fifth IEEE international conference on data mining, pp 74–81
Borgwardt KM, Ong CS, Schönauer S, Vishwanathan SVN, Smola AJ, Kriegel HP (2005) Protein function prediction via graph kernels. Bioinformatics 21(Suppl 1):i47–i56
Bruna J, Zaremba W, Szlam A, LeCun Y (2014) Spectral networks and deep locally connected networks on graphs. In: International conference on learning representations
Chang CC, Lin CJ (2011) LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2:27:1–27:27. Software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm
Defferrard M, Bresson X, Vandergheynst P (2016) Convolutional neural networks on graphs with fast localized spectral filtering. In: Advances in neural information processing systems, pp 3844–3852
Duvenaud DK, Maclaurin D, Iparraguirre J, Bombarell R, Hirzel T, Aspuru-Guzik A, Adams RP (2015) Convolutional networks on graphs for learning molecular fingerprints. In: Advances in neural information processing systems, pp 2224–2232
Feragen A, Kasenburg N, Petersen J, Bruijne MD, Borgwardt K (2013) Scalable kernels for graphs with continuous attributes. In: Burges C, Bottou L, Welling M, Ghahramani Z, Weinberger K (eds) Advances in neural information processing systems, pp 216–224. Erratum available at http://image.diku.dk/aasa/papers/graphkernels_nips_erratum.pdf
Fout A, Byrd J, Shariat B, Ben-Hur A (2017) Protein interface prediction using graph convolutional networks. In: Advances in neural information processing systems, pp 6533–6542
Fröhlich H, Wegner JK, Sieker F, Zell A (2005) Optimal assignment kernels for attributed molecular graphs. In: Proceedings of the 22nd international conference on machine learning. ACM, New York, NY, USA, ICML ’05, pp 225–232
Gärtner T, Flach P, Wrobel S (2003) On graph kernels: hardness results and efficient alternatives. In: Learning theory and kernel machines, Lecture Notes in Computer Science, vol 2777. Springer, pp 129–143
Ghosh S, Das N, Gonçalves T, Quaresma P, Kundu M (2018) The journey of graph kernels through two decades. Comput Sci Rev 27:88–111
Gilmer J, Schoenholz SS, Riley PF, Vinyals O, Dahl GE (2017) Neural message passing for quantum chemistry. In: 33rd International conference on machine learning
Hamilton WL, Ying R, Leskovec J (2017a) Inductive representation learning on large graphs. In: Advances in neural information processing systems, pp 1025–1035
Hamilton WL, Ying R, Leskovec J (2017b) Representation learning on graphs: methods and applications. IEEE Data Eng Bull 40(3):52–74
Harchaoui Z, Bach F (2007) Image classification with segmentation graph kernels. In: IEEE conference on computer vision and pattern recognition
Haussler D (1999) Convolution kernels on discrete structures. Tech. Rep. UCSC-CRL-99-10, University of California, Santa Cruz, CA, USA
Hido S, Kashima H (2009) A linear-time graph kernel. In: The ninth IEEE international conference on data mining, pp 179–188
Horváth T, Gärtner T, Wrobel S (2004) Cyclic pattern kernels for predictive graph mining. In: Proceedings of the twelfth ACM SIGKDD international conference on knowledge discovery and data mining, pp 158–167
Joachims T (2006) Training linear SVMs in linear time. In: Proceedings of the twelfth ACM SIGKDD international conference on knowledge discovery and data mining, pp 217–226
Johansson FD, Dubhashi D (2015) Learning with similarity functions on graphs using matchings of geometric embeddings. In: Proceedings of the 21st ACM SIGKDD international conference on knowledge discovery and data mining. ACM, New York, NY, USA, KDD ’15, pp 467–476
Kang U, Tong H, Sun J (2012) Fast random walk graph kernel. In: Proceedings of the 2012 SIAM international conference on data mining, pp 828–838
Kar P, Karnick H (2012) Random feature maps for dot product kernels. In: Proceedings of the fifteenth international conference on artificial intelligence and statistics, AISTATS 2012, La Palma, Canary Islands, April 21–23, 2012, pp 583–591
Kashima H, Tsuda K, Inokuchi A (2003) Marginalized kernels between labeled graphs. In: Proceedings of the twentieth international conference on machine learning, pp 321–328
Kersting K, Kriege NM, Morris C, Mutzel P, Neumann M (2016) Benchmark data sets for graph kernels. http://graphkernels.cs.tu-dortmund.de
Kipf TN, Welling M (2017) Semi-supervised classification with graph convolutional networks. In: International conference on learning representations
Kondor R, Pan H (2016) The multiscale Laplacian graph kernel. In: Advances in neural information processing systems, pp 2982–2990
Kriege N, Mutzel P (2012) Subgraph matching kernels for attributed graphs. In: Proceedings of the 29th international conference on machine learning. http://www.icml.cc/Omnipress
Kriege N, Neumann M, Kersting K, Mutzel M (2014) Explicit versus implicit graph feature maps: a computational phase transition for walk kernels. In: 2014 IEEE international conference on data mining, pp 881–886
Kriege NM, Giscard PL, Wilson R (2016) On valid optimal assignment kernels and applications to graph classification. In: Advances in neural information processing systems. Curran Associates, Inc., pp 1623–1631
Kriege NM, Johansson FD, Morris C (2019) A survey on graph kernels. CoRR. arXiv:1903.11835
Mahé P, Ueda N, Akutsu T, Perret JL, Vert JP (2004) Extensions of marginalized graph kernels. In: Proceedings of the twenty-first international conference on machine learning, p 70
Martino GDS, Navarin N, Sperduti A (2012) A tree-based kernel for graphs. In: Proceedings of the 2012 SIAM international conference on data mining. SIAM/Omnipress, pp 975–986
Martino GDS, Navarin N, Sperduti A (2018) Tree-based kernel for graphs with continuous attributes. IEEE Trans Neural Netw Learn Syst 29(7):3270–3276
Merkwirth C, Lengauer T (2005) Automatic generation of complementary descriptors with molecular graph networks. J Chem Inf Model 45(5):1159–1168
Morris C, Kriege NM, Kersting K, Mutzel P (2016) Faster kernels for graphs with continuous attributes via hashing. In: Bonchi F, Domingo-Ferrer J (eds) IEEE international conference on data mining (ICDM)
Narayanan A, Chandramohan M, Chen L, Liu Y, Saminathan S (2016) subgraph2vec: learning distributed representations of rooted sub-graphs from large graphs. In: Workshop on mining and learning with graphs. arXiv:1606.08928
Neumann M, Garnett R, Bauckhage C, Kersting K (2016) Propagation kernels: efficient graph kernels from propagated information. Mach Learn 102(2):209–245
Nikolentzos G, Meladianos P, Vazirgiannis M (2017) Matching node embeddings for graph similarity. In: AAAI. AAAI Press, pp 2429–2435
Nikolentzos G, Meladianos P, Limnios S, Vazirgiannis M (2018) A degeneracy framework for graph similarity. In: IJCAI, pp 2595–2601. http://www.ijcai.org
Orsini F, Frasconi P, De Raedt L (2015) Graph invariant kernels. In: Proceedings of the twenty-fourth international joint conference on artificial intelligence, pp 3756–3762
Pham N, Pagh R (2013) Fast and scalable polynomial kernels via explicit feature maps. In: Proceedings of the 19th ACM SIGKDD international conference on knowledge discovery and data mining. ACM, New York, NY, USA, KDD ’13, pp 239–247. https://doi.org/10.1145/2487575.2487591
Rahimi A, Recht B (2008) Random features for large-scale kernel machines. In: Advances in neural information processing systems, pp 1177–1184
Ramon J, Gärtner T (2003) Expressivity versus efficiency of graph kernels. In: First international workshop on mining graphs, trees and sequences
Scarselli F, Gori M, Tsoi AC, Hagenbuchner M, Monfardini G (2009) The graph neural network model. Trans Neural Netw 20(1):61–80
Schiavinato M, Gasparetto A, Torsello A (2015) Transitive assignment kernels for structural classification. In: Feragen A, Pelillo M, Loog M (eds) Similarity-based pattern recognition: third international workshop, SIMBAD 2015, Copenhagen, Denmark, October 12–14, 2015. Springer International Publishing, Cham, pp 146–159
Schütt K, Kindermans PJ, Sauceda HE, Chmiela S, Tkatchenko A, Müller KR (2017) SchNet: a continuous-filter convolutional neural network for modeling quantum interactions. In: Advances in neural information processing systems, pp 992–1002
Shawe-Taylor J, Cristianini N (2004) Kernel methods for pattern analysis. Cambridge University Press, New York
Shervashidze N, Borgwardt K (2009) Fast subtree kernels on graphs. In: Bengio Y, Schuurmans D, Lafferty J, Williams CKI, Culotta A (eds) Advances in neural information processing systems, vol 22, pp 1660–1668
Shervashidze N, Vishwanathan S, Petri TH, Mehlhorn K, Borgwardt KM (2009) Efficient graphlet kernels for large graph comparison. In: 12th International conference on artificial intelligence and statistics
Shervashidze N, Schweitzer P, van Leeuwen EJ, Mehlhorn K, Borgwardt KM (2011) Weisfeiler-Lehman graph kernels. J Mach Learn Res 12:2539–2561
Shi Q, Petterson J, Dror G, Langford J, Smola A, Vishwanathan S (2009) Hash kernels for structured data. J Mach Learn Res 10:2615–2637
Shin K, Kuboyama T (2010) A generalization of Haussler’s convolution kernel—mapping kernel and its application to tree kernels. J Comput Sci Technol 25:1040–1054
Su Y, Han F, Harang RE, Yan X (2016) A fast kernel for attributed graphs. In: Proceedings of the 2016 SIAM international conference on data mining
Sugiyama M, Borgwardt KM (2015) Halting in random walk kernels. In: Advances in neural information processing systems, pp 1630–1638
Swamidass SJ, Chen J, Bruand J, Phung P, Ralaivola L, Baldi P (2005) Kernels for small molecules and the prediction of mutagenicity, toxicity and anti-cancer activity. Bioinformatics 21(Suppl 1):i359–i368. https://doi.org/10.1093/bioinformatics/bti1055
Vedaldi A, Zisserman A (2012) Efficient additive kernels via explicit feature maps. IEEE Trans Pattern Anal Mach Intell 34(3):480–492
Vert JP (2008) The optimal assignment kernel is not positive definite. CoRR abs/0801.4061
Vishwanathan SVN, Schraudolph NN, Kondor RI, Borgwardt KM (2010) Graph kernels. J Mach Learn Res 11:1201–1242
Wale N, Watson IA, Karypis G (2008) Comparison of descriptor spaces for chemical compound retrieval and classification. Knowl Inf Syst 14(3):347–375
Yanardag P, Vishwanathan SVN (2015) Deep graph kernels. In: 21st ACM SIGKDD international conference on knowledge discovery and data mining, pp 1365–1374
Ying R, He R, Chen K, Eksombatchai P, Hamilton WL, Leskovec J (2018a) Graph convolutional neural networks for web-scale recommender systems. In: ACM SIGKDD international conference on knowledge discovery & data mining
Ying R, You J, Morris C, Ren X, Hamilton WL, Leskovec J (2018b) Hierarchical graph representation learning with differentiable pooling. In: Advances in neural information processing systems
Zhang M, Cui Z, Neumann M, Chen Y (2018a) An end-to-end deep learning architecture for graph classification. In: AAAI conference on artificial intelligence
Zhang Y, Wang L, Wang L (2018b) A comprehensive evaluation of graph kernels for unattributed graphs. Entropy 20(12):984
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Hanghang Tong.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A preliminary version of this paper appeared in the proceedings of the IEEE International Conference on Data Mining (ICDM) in 2014 (Kriege et al. 2014).
This work has been supported by the Deutsche Forschungsgemeinschaft (DFG) within the Collaborative Research Center SFB 876 “Providing Information by Resource-Constrained Data Analysis”, project A6 “Resource-efficient Graph Mining”.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kriege, N.M., Neumann, M., Morris, C. et al. A unifying view of explicit and implicit feature maps of graph kernels. Data Min Knowl Disc 33, 1505–1547 (2019). https://doi.org/10.1007/s10618-019-00652-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10618-019-00652-0