Abstract
Incidents of food fraud have occurred worldwide, particularly in the form of meat adulteration. In this study, molecular probes were developed using the Random amplification of polymorphic DNA (RAPD) polymerase chain reaction (PCR) technique in order to identify three beef subspecies–Holstein, Angus, and Taiwan Yellow Cattle. Four RAPD-PCR 10-nucleotide primers were chosen out of a total of 60 primers. The selection was based on the reproducibility of species-specific amplicons able to detect various origins of cattle breeds. The results demonstrated that primer OPK12 produced three unique amplicons (1100 bp, 1000 bp and 480 bp) in Holstein; primer OPK14 generated one amplicon that only appeared in Holstein and Angus (200 bp); primer OPK19 amplified two species-specific amplicons in Holstein measuring 550 bp and 650 bp, respectively. However, due to the relatively lower repeatability of RAPD-PCR, higher and more specific testing repeats were required to increase the accuracy of the conclusion.


Similar content being viewed by others
References
Abdel-Rahman SM, Ahmed MMM. Rapid and sensitive identification of buffalo’s, cattle’s and sheep’s milk using species-specific PCR and PCR–RFLP techniques. Food Control. 18: 1246–1249 (2007)
Chen S, Zhang Y, Li H, Wang J, Chen W, Zhou Y, Zhou S. Differentiation of fish species in Taiwan Strait by PCR-RFLP and lab-on-a-chip system. Food Control. 44: 26–34 (2014)
Combes MC, Joët T, Lasherme P. Development of a rapid and efficient DNA-based method to detect and quantify adulterations in coffee (Arabica versus Robusta). Food Control. 88: 198–206 (2018)
Fajardo V, Gonzlez I, Martn I, Rojas M, Hernndez PE, Garca T, Martn R. Real-time PCR for quantitative detection of chamois (Rupicapra rupicapra) and pyrenean ibex (Capra pyrenaica) in meat mixtures. J. AOAC Int. 91: 103–111 (2008)
Kaur J, Lee S, Park YS, Sharma A. RAPD analysis of Leuconostoc mesenteroides strains associated with vegetables and food products from Korea. LWT-Food Sci. Technol. 77: 383–388 (2017)
Keeratipibul S, Techaruwichit P. Tracking sources of Listeria contamination in a cooked chicken meat factory by PCR-RAPD-based DNA fingerprinting. Food Control. 27: 64–72 (2012)
Kim MJ, Kim HY. Species identification of commercial jerky products in food and feed using direct pentaplex PCR assay. Food Control. 78: 1–6 (2017)
Kim MJ, Kim HY. Development of a fast duplex real-time PCR assay for simultaneous detection of chicken and pigeon in raw and heat-treated. Food Control. 85: 1–5 (2018)
Kitpipit T, Sittichan K, Thanakiatkrai P. Direct-multiplex PCR assay for meat species identification in food products. Food Chem. 163: 77–82 (2014)
Koh MC, Lim CH, Chua SB, Chew ST, Phang STW. Random amplified polymorphic DNA (RAPD) fingerprints for identification of red meat animal species. Meat Sci. 48: 275–285 (1998)
Kumar A, Kumar RR, Sharma BD, Gokulakrishnan P, Mendiratta SK & Sharma D. Identification of Species Origin of Meat and Meat Products on the DNA Basis: a review. Crit. Rev. Food Sci. 55: 1340–1351 (2015)
Kwon GH, Lee HA, Park JY, Kim JS, Lim J, Park CK, Kwon DY, Kim YS, Kim JH. Development of a RAPD-PCR method for identification of Bacillus species isolated from Cheonggukjang. Int. J. Food Microbol. 129: 282–287 (2009)
Martinez I, Yman IM. Species identification in meat products by RAPD analysis. Food Res. Int. 31: 459–466 (1998)
Peng GJ, Chang MH, Fang M, Liao CD, Tsai CF, Tseng SH, Kao YM, Chou HK, Cheng HF. Incidents of major food adulteration in Taiwan between 2011 and 2015. Food Control. 72: 145–152 (2017)
Perez T, Albornoz J, Dominguez A. An evaluation of RAPD fragment reproducibility and nature. Mol. Ecol. 7: 1347–1357 (1998)
Rahman MM, Ali ME, Hamid SBA, Bhassu S, Mustafa S, Al Amin M, Razzak MA. Lab-on-a-Chip PCR-RFLP Assay for the Detection of Canine DNA in Burger Formulations. Food Anal. Methods. 8: 1598–1606 (2015)
Rahmati S, Julkapli NM, Yehye WA, Basirun WJ. Identification of meat origin in food products–A review. Food Control. 68: 379–390 (2016)
Rastogi G, Dharne MS, Walujkar S, Kumar A, Patole MS, Shouche YS. Species identification and authentication of tissues of animal origin using mitochondrial and nuclear markers. Meat Sci. 76: 666–674 (2007)
Ropodi AI, Panagou EZ, Nychas GJE. Multispectral imaging (MSI): A promising method for the detection of minced beef adulteration with horsemeat. Food Control. 73: 57–63 (2017)
Shehata MM. The Roles of Seed Proteins and RAPD-PCR in Genotyping Variabilities of Some Wheat (Triticum vulgare L.) Cultivars. Pakistan J. Biol. Sci. 7: 984–994 (2004)
Soares S, Amaral JS, Oliveira MBP, Mafra I. A SYBR Green real-time PCR assay to detect and quantify pork meat in processed poultry meat products. Meat Sci. 94: 115–120 (2013)
Sobrino-Gregorio L, Vilanova S, Prohens J, Escriche I. Detection of honey adulteration by conventional and real-time PCR. Food Control. 95: 57–62 (2018)
Thanakiatkrai P, Dechnakarin J, Ngasaman R, Kitpipit T. Direct pentaplex PCR assay: An adjunct panel for meat species identification in Asian food products. Food Chem. 271: 762–772 (2019)
Velioglu HM, Sezer B, Bilge G, Baytur SE, Boyaci IH. Identification of offal adulteration in beef by laser induced breakdown spectroscopy (LIBS). Meat Sci. 138: 28–33 (2018)
Xu R, Wei S, Zhou G, Ren J, Liu Z, Tang S, Cheung PCK, Wu X. Multiplex TaqMan locked nucleic acid real-time PCR for the differential identification of various meat and meat products. Meat Sci. 137: 41–46 (2018)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
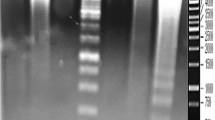
Results of PCR fingerprinting of false detection. H: Holstein, A: Angus, Y: Taiwan Yellow Cattle, HAY: Holstein + Angus + Taiwan Yellow Cattle. M: 100 bp Plus DNA ladder, lane 1: unique amplicons (1000 bp, 1100 bp and 480 bp) in Holstein by primer OPK12 (shown in arrow mark), lane 2: Angus with primer OPK12, lane 3: Taiwan Yellow Cattle with primer OPK12
Results of PCR fingerprinting regarding false detection. Amplicon showed only in Holstein and Angus. H: Holstein, A: Angus, Y: Taiwan Yellow Cattle, HAY: Holstein + Angus + Taiwan Yellow Cattle. M: 100 bp Plus DNA ladder, lane 1: amplicon with 180 bp molecular weights in Holstein by primer OPK14 (shown in arrow mark), lane 2: amplicon with 180 bp molecular weights in Angus by primer OPK14 (shown in arrow mark), lane 3: Taiwan Yellow Cattle with primer OPK14
Results of PCR fingerprinting regarding false detection. H: Holstein, A: Angus, Y: Taiwan Yellow Cattle, HAY: Holstein + Angus + Taiwan Yellow Cattle. M: 100 bp Plus DNA ladder, lane 1: unique amplicons (550 bp, 650 bp) in Holstein by primer OPK19 (shown in arrow mark), lane 2 Angus with primer OPK19, lane 3: Taiwan Yellow Cattle with primer OPK19
Rights and permissions
About this article
Cite this article
Lin, CC., Tang, PC. & Chiang, HI. Development of RAPD-PCR assay for identifying Holstein, Angus, and Taiwan Yellow Cattle for meat adulteration detection. Food Sci Biotechnol 28, 1769–1777 (2019). https://doi.org/10.1007/s10068-019-00607-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10068-019-00607-7