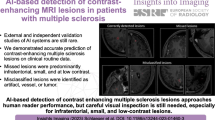
Abstract
Objectives
Patients with multiple sclerosis (MS) regularly undergo MRI for assessment of disease burden. However, interpretation may be time consuming and prone to intra- and interobserver variability. Here, we evaluate the potential of artificial neural networks (ANN) for automated volumetric assessment of MS disease burden and activity on MRI.
Methods
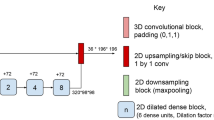
A single-institutional dataset with 334 MS patients (334 MRI exams) was used to develop and train an ANN for automated identification and volumetric segmentation of T2/FLAIR-hyperintense and contrast-enhancing (CE) lesions. Independent testing was performed in a single-institutional longitudinal dataset with 82 patients (266 MRI exams). We evaluated lesion detection performance (F1 scores), lesion segmentation agreement (DICE coefficients), and lesion volume agreement (concordance correlation coefficients [CCC]). Independent evaluation was performed on the public ISBI-2015 challenge dataset.
Results
The F1 score was maximized in the training set at a detection threshold of 7 mm3 for T2/FLAIR lesions and 14 mm3 for CE lesions. In the training set, mean F1 scores were 0.867 for T2/FLAIR lesions and 0.636 for CE lesions, as compared to 0.878 for T2/FLAIR lesions and 0.715 for CE lesions in the test set. Using these thresholds, the ANN yielded mean DICE coefficients of 0.834 and 0.878 for segmentation of T2/FLAIR and CE lesions in the training set (fivefold cross-validation). Corresponding DICE coefficients in the test set were 0.846 for T2/FLAIR lesions and 0.908 for CE lesions, and the CCC was ≥ 0.960 in each dataset.
Conclusions
Our results highlight the capability of ANN for quantitative state-of-the-art assessment of volumetric lesion load on MRI and potentially enable a more accurate assessment of disease burden in patients with MS.
Key Points
• Artificial neural networks (ANN) can accurately detect and segment both T2/FLAIR and contrast-enhancing MS lesions in MRI data.
• Performance of the ANN was consistent in a clinically derived dataset, with patients presenting all possible disease stages in MRI scans acquired from standard clinical routine rather than with high-quality research sequences.
• Computer-aided evaluation of MS with ANN could streamline both clinical and research procedures in the volumetric assessment of MS disease burden as well as in lesion detection.



Similar content being viewed by others
Abbreviations
- ANN:
-
Artificial neural networks
- CCC:
-
Concordance correlation coefficients
- CE:
-
Contrast enhancing
- FLAIR:
-
Fluid-attenuated inversion recovery
- GT:
-
Ground truth
- LPPV:
-
Lesion-wise positive predictive value
- LTPR:
-
Lesion-wise true positive rate
- MRI:
-
Magnetic resonance imaging
- MS:
-
Multiple sclerosis
- PD-w:
-
Proton density weighted
- T2-w:
-
T2 weighted
References
Filippi M, Bar-Or A, Piehl F et al (2018) Multiple sclerosis. Nat Rev Dis Primers 4:43
Sweeney EM, Shinohara RT, Shiee N et al (2013) OASIS is automated statistical inference for segmentation, with applications to multiple sclerosis lesion segmentation in MRI. Neuroimage Clin 2:402–413
Carass A, Roy S, Jog A et al (2017) Longitudinal multiple sclerosis lesion segmentation: resource and challenge. Neuroimage 148:77–102
Jesson A, Arbel T (2015) Hierarchical MRF and random forest segmentation of MS lesions and healthy tissues in brain MRI. Proceedings of the 2015 Longitudinal Multiple Sclerosis Lesion Segmentation Challenge:1–2
Geremia E, Clatz O, Menze BH, Konukoglu E, Criminisi A, Ayache N (2011) Spatial decision forests for MS lesion segmentation in multi-channel magnetic resonance images. Neuroimage 57:378–390
Fartaria MJ, Bonnier G, Roche A et al (2016) Automated detection of white matter and cortical lesions in early stages of multiple sclerosis. J Magn Reson Imaging 43:1445–1454
Commowick O, Cervenansky F, Ameli R (2016) Proceedings of the 1st MSSEG challenge on multiple sclerosis lesions segmentation challenge using a data managementand processing infrastructure. MICCAI-MSSEG
Danelakis A, Theoharis T, Verganelakis DA (2018) Survey of automated multiple sclerosis lesion segmentation techniques on magnetic resonance imaging. Comput Med Imaging Graph 70:83–100
Aslani S, Dayan M, Storelli L et al (2018) Multi-branch convolutional neural network for multiple sclerosis lesion segmentation. arXiv:1811.02942
Brosch T, Tang LY, Yoo Y, Li DK, Traboulsee A, Tam R (2016) Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans Med Imaging 35:1229–1239
Andermatt S, Pezold S, Cattin PC (2017) Automated segmentation of multiple sclerosis lesions using multi-dimensional gated recurrent units. In: Crimi A, Bakas S, Kuijf H, Menze B, Reyes M (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2017. Lecture Notes in Computer Science, vol 10670. Springer, Cham
Ghafoorian M, Platel B (2015) Convolutional neural networks for ms lesion segmentation, method description of diag team. Proceedings of the 2015 Longitudinal Multiple Sclerosis Lesion Segmentation Challenge:1–2
Valverde S, Cabezas M, Roura E et al (2017) Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. Neuroimage 155:159–168
Brosch T, Tang LY, Youngjin Y, Li DK, Traboulsee A, Tam R (2016) Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans Med Imaging 35:1229–1239
Aslani S, Dayan M, Storelli L et al (2019) Multi-branch convolutional neural network for multiple sclerosis lesion segmentation. Neuroimage 196:1–15
Saindane AM (2019) Is gadolinium-based contrast material needed for MRI follow-up of multiple sclerosis? Radiology 291:436–437
Kaunzner UW, Gauthier SA (2017) MRI in the assessment and monitoring of multiple sclerosis: an update on best practice. Ther Adv Neurol Disord 10:247–261
Polman CH, Reingold SC, Banwell B et al (2011) Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 69:292–302
Kickingereder P, Isensee F, Tursunova I et al (2019) Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. Lancet Oncol 20:728–740
Yushkevich PA, Piven J, Hazlett HC et al (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31:1116–1128
Isensee F, Petersen J, Klein A et al (2018) nnU-Net: self-adapting framework for U-Net-based medical image segmentation. arXiv:1809.10486
Simpson AL, Antonelli M, Bakas S et al (2019) A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv:1902.09063
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Springer, Cham
Roy S, Butman JA, Reich DS, Calabresi PA, Pham DL (2018) Multiple sclerosis lesion segmentation from brain MRI via fully convolutional neural networks. arXiv:1803.09172
Marcus DS, Olsen TR, Ramaratnam M, Buckner RL (2007) The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics 5:11–34
Thompson AJ, Banwell BL, Barkhof F et al (2018) Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol 17:162–173
Eichinger P, Schon S, Pongratz V et al (2019) Accuracy of unenhanced MRI in the detection of new brain lesions in multiple sclerosis. Radiology 291:429–435
Funding
Dr. Kickingereder, MBA was supported by the Medical Faculty Heidelberg Postdoc-Program and the Else Kröner-Fresenius Foundation (Else-Kröner Memorial Scholarship).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Dr. Philipp Kickingereder, MBA.
Conflict of interest
The authors declare that they have no competing interests.
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained at the local ethics committee of the medical faculty of the University of Heidelberg.
Methodology
• Retrospective
• Diagnostic or prognostic study
• Performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 294 kb)
Rights and permissions
About this article
Cite this article
Brugnara, G., Isensee, F., Neuberger, U. et al. Automated volumetric assessment with artificial neural networks might enable a more accurate assessment of disease burden in patients with multiple sclerosis. Eur Radiol 30, 2356–2364 (2020). https://doi.org/10.1007/s00330-019-06593-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-019-06593-y