Abstract
Objectives
To perform test-retest reproducibility analyses for deep learning–based automatic detection algorithm (DLAD) using two stationary chest radiographs (CRs) with short-term intervals, to analyze influential factors on test-retest variations, and to investigate the robustness of DLAD to simulated post-processing and positional changes.
Methods
This retrospective study included patients with pulmonary nodules resected in 2017. Preoperative CRs without interval changes were used. Test-retest reproducibility was analyzed in terms of median differences of abnormality scores, intraclass correlation coefficients (ICC), and 95% limits of agreement (LoA). Factors associated with test-retest variation were investigated using univariable and multivariable analyses. Shifts in classification between the two CRs were analyzed using pre-determined cutoffs. Radiograph post-processing (blurring and sharpening) and positional changes (translations in x- and y-axes, rotation, and shearing) were simulated and agreement of abnormality scores between the original and simulated CRs was investigated.
Results
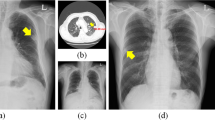
Our study analyzed 169 patients (median age, 65 years; 91 men). The median difference of abnormality scores was 1–2% and ICC ranged from 0.83 to 0.90. The 95% LoA was approximately ± 30%. Test-retest variation was negatively associated with solid portion size (β, − 0.50; p = 0.008) and good nodule conspicuity (β, − 0.94; p < 0.001). A small fraction (15/169) showed discordant classifications when the high-specificity cutoff (46%) was applied to the model outputs (p = 0.04). DLAD was robust to the simulated positional change (ICC, 0.984, 0.996), but relatively less robust to post-processing (ICC, 0.872, 0.968).
Conclusions
DLAD was robust to the test-retest variation. However, inconspicuous nodules may cause fluctuations of the model output and subsequent misclassifications.
Key Points
• The deep learning–based automatic detection algorithm was robust to the test-retest variation of the chest radiographs in general.
• The test-retest variation was negatively associated with solid portion size and good nodule conspicuity.
• High-specificity cutoff (46%) resulted in discordant classifications of 8.9% (15/169; p = 0.04) between the test-retest radiographs.





Similar content being viewed by others
Abbreviations
- CI:
-
Confidence interval
- CR:
-
Chest radiograph
- Diffscore :
-
Difference of abnormality scores between the test-retest chest radiographs
- DLAD:
-
Deep learning–based automatic detection algorithm
- ICC:
-
Intraclass correlation coefficient
- IQR:
-
Interquartile range
- LoA:
-
Limits of agreement
References
Hwang EJ, Park S, Jin KN et al (2019) Development and validation of a deep learning based automated detection algorithm for major thoracic diseases on chest radiographs. JAMA Netw Open 2:e191095
Nam JG, Park S, Hwang EJ et al (2019) Development and validation of deep learning-based automatic detection algorithm for malignant pulmonary nodules on chest radiographs. Radiology 290:218–228
Annarumma M, Withey SJ, Bakewell RJ, Pesce E, Goh V, Montana G (2019) Automated triaging of adult chest radiographs with deep artificial neural networks. Radiology 291:196–202
Dunnmon JA, Yi D, Langlotz CP, Re C, Rubin DL, Lungren MP (2019) Assessment of convolutional neural networks for automated classification of chest radiographs. Radiology 290:537–544
Lakhani P, Sundaram B (2017) Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 284:574–582
Hwang EJ, Park S, Jin KN et al (2019) Development and validation of a deep learning-based automatic detection algorithm for active pulmonary tuberculosis on chest radiographs. Clin Infect Dis 69:739–747
Rajpurkar P, Irvin J, Ball RL et al (2018) Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med 15:e1002686
Singh R, Kalra MK, Nitiwarangkul C et al (2018) Deep learning in chest radiography: detection of findings and presence of change. PLoS One 13:e0204155
Taylor AG, Mielke C, Mongan J (2018) Automated detection of moderate and large pneumothorax on frontal chest X-rays using deep convolutional neural networks: a retrospective study. PLoS Med 15:e1002697
Sullivan DC, Obuchowski NA, Kessler LG et al (2015) Metrology standards for quantitative imaging biomarkers. Radiology 277:813–825
Kim H, Goo JM, Ohno Y et al (2019) Effect of reconstruction parameters on the quantitative analysis of chest computed tomography. J Thorac Imaging 34:92–102
Kim H, Park CM, Gwak J et al (2019) Effect of CT reconstruction algorithm on the diagnostic performance of radiomics models: a task-based approach for pulmonary subsolid nodules. AJR Am J Roentgenol 212:505–512
Kim H, Park CM, Hwang EJ, Ahn SY, Goo JM (2018) Pulmonary subsolid nodules: value of semi-automatic measurement in diagnostic accuracy, diagnostic reproducibility and nodule classification agreement. Eur Radiol 28:2124–2133
de Lacey G, Morley S, Berman L (2007) The chest x-ray: a survival guide, 1st edn. Saunders, Elsevier, Amsterdam
Sedgwick P (2013) Limits of agreement (Bland-Altman method). BMJ 346:f1630
Bland JM, Altman DG (1999) Measuring agreement in method comparison studies. Stat Methods Med Res 8:135–160
Koo TK, Li MY (2016) A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med 15:155–163
Berenguer R, Pastor-Juan MDR, Canales-Vazquez J et al (2018) Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology 288:407–415
Irvin J, Rajpurkar P, Ko M et al (2019) Chexpert: a large chest radiograph dataset with uncertainty labels and expert comparison. arXiv:1901.07031 available via https://arxiv.org/abs/1901.07031.
He L, Huang Y, Ma Z, Liang C, Liang C, Liu Z (2016) Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep 6:34921
Li Y, Lu L, Xiao M et al (2018) CT slice thickness and convolution kernel affect performance of a radiomic model for predicting EGFR status in non-small cell lung cancer: a preliminary study. Sci Rep 8:17913
Park SH (2019) Diagnostic case-control versus diagnostic cohort studies for clinical validation of artificial intelligence algorithm performance. Radiology 290:272–273
Goodfellow I, McDaniel P, Papernot N (2018) Making machine learning robust against adversarial inputs. Commun ACM 61:56–66
Funding
This study was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT & Future Planning (grant number 2017R1A2B4008517).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Chang Min Park.
Conflict of interest
The authors (H.K., C.M.P., and J.M.G.) received research grants from Lunit Inc. (Seoul, South Korea), which developed the deep learning–based detection algorithm (Lunit INISIGHT for Chest Radiography) used in this study. However, Lunit Inc. had no role in the study design; in the collection, analysis, and interpretation of the data; in the writing of the report; and in the decision to submit the article for publication.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 15 kb)
Rights and permissions
About this article
Cite this article
Kim, H., Park, C.M. & Goo, J.M. Test-retest reproducibility of a deep learning–based automatic detection algorithm for the chest radiograph. Eur Radiol 30, 2346–2355 (2020). https://doi.org/10.1007/s00330-019-06589-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-019-06589-8