Abstract
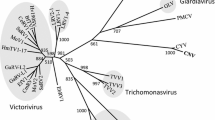
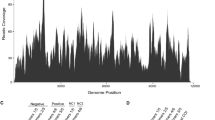
A new polycipivirus was identified in the arboreal ant Colobopsis shohki. The viral RNA was 11,855 nt in length with five 5’-proximal open reading frames (ORFs) encoding structural proteins and a long 3’ ORF encoding the replication polyprotein. The protein sequences of these ORFs had significant similarity to those of the polycipiviruses Lasius niger virus 1 and Solenopsis invicta virus 2. The results of phylogenetic analysis and its genome organization suggested that this virus belongs to the genus Sopolycivirus in the family Polycipiviridae. The name “Colobopsis shohki virus 1” (CshV1) is proposed for the new virus.


Similar content being viewed by others
References
Olendraite I, Brown K., Valles SM, Firth AE, Chen Y, Guérin DMA, Hashimoto Y, Herrero S, de Miranda JR, Ryabov E, ICTV Report Consortium (2019) ICTV virus taxonomy profile: Polycipiviridae. J Gen Virol 100:554–555
Olendraite I, Lukhovitskaya NI, Porter SD, Valles SM, Firth AE (2017) Polycipiviridae: a proposed new family of polycistronic picorna-like RNA viruses. J Gen Virol 98:2368–2378
Koyama S, Urayama S, Ohmatsu T, Sassa Y, Sakai C, Takata M, Hayashi S, Nagai M, Furuya T, Moriyama H, Satoh T, Ono S, Mizutani T (2015) Identification, characterization and full-length sequence analysis of a novel dsRNA virus isolated from the arboreal ant Camponotus yamaokai. J Gen Virol 96:1930–1937
Koyama S, Sakai C, Thomas CE, Nunoura T, Urayama S (2016) A new member of the family Totiviridae associated with arboreal ants (Camponotus nipponicus). Arch Virol 161:2043–2045
Urayama S, Yoshida-Takashima Y, Yoshida M, Tomaru Y, Moriyama H, Takai K, Nunoura T (2015) A new fractionation and recovery method of viral genomes based on nucleic acid composition and structure using tandem column chromatography. Microbes Environ 30:199–203
Urayama S, Takaki Y, Nunoura T (2016) FLDS: a comprehensive dsRNA sequencing method for intracellular RNA virus surveillance. Microbes Environ 31:33–40
Urayama S, Takaki Y, Nishi S, Yoshida-Takashima Y, Deguchi S, Takai K, Nunoura T (2018) Unveiling the RNA virosphere associated with marine microorganisms. Mol Ecol Resour 18:1444–1455
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. Journal 17:10–12
Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864
Zimmermann L, Stephens A, Nam SZ, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V (2018) A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 430:2237–2243
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PD (2000) The protein data bank. Nucleic Acids Res 28:235–242
Hashimoto Y, Valles SM (2008) Infection characteristics of Solenopsis invicta virus 2 in the red imported fire ant, Solenopsis invicta. J Invertebr Pathol 99:136–140
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Acknowledgements
This research was supported in part by a Grant-in-Aid for Challenging Research (Pioneering) from JSPS (18H05368) and a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Science, Sports, and Technology (MEXT) of Japan (nos. 16H06429, 16K21723, and 16H06437). JAMSTEC has filed a patent application related to FLDS, of which S.U., T.N. are the named inventors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Handling Editor: T. K. Frey.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fukasawa, F., Hirai, M., Takaki, Y. et al. A new polycipivirus identified in Colobopsis shohki. Arch Virol 165, 761–763 (2020). https://doi.org/10.1007/s00705-019-04510-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04510-8