Abstract
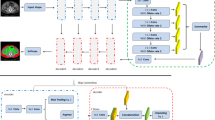
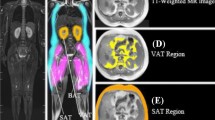
One major role of an accurate distribution of abdominal adipose tissue is to predict disease risk. This paper proposes a novel effective three-level convolutional neural network (CNN) approach to automate the selection of abdominal computed tomography (CT) images on large-scale CT scans and automatically quantify the visceral and subcutaneous adipose tissue. First, the proposed framework employs support vector machine (SVM) classifier with a configured parameter to cluster abdominal CT images from screening patients. Second, a pyramid dilation network (DilaLab) is designed based on CNN, to address the complex distribution and non-abdominal internal adipose tissue problems of biomedical image segmentation in visceral adipose tissue. Finally, since the trained DilaLab implicitly encodes the fat-related learning, the transferred DilaLab learning and a simple decoder constitute a new network (DilaLabPlus) for quantifying subcutaneous adipose tissue. The networks are trained not only all available CT images but also with a limited number of CT scans, such as 70 samples including a 10% validation subset. All networks are yielding more precise results. The mean accuracy of the configured SVM classifier yields promising performance of 99.83%, while DilaLabPlus achieves a remarkable performance improvement an with average of 98.08 ± 0.84% standard deviation and 0.7 ± 0.8% standard deviation false-positive rate. The performance of DilaLab yields average 97.82 ± 1.34% standard deviation and 1.23 ± 1.33% standard deviation false-positive rate. This study demonstrates considerable improvement in feasibility and reliability for the fully automated recognition of abdominal CT slices and segmentation of selected abdominal CT in subcutaneous and visceral adipose tissue, and it has a high agreement with a manually annotated biomarker.









Similar content being viewed by others
References
Agarwal, C., A. H. Dallal, M. R. Arbabshirani, A. Patel, and G. Moore, Unsupervised quantification of abdominal fat from CT images using greedy snakes. In: Society of Photo-optical Instrumentation Engineers, 2017, p. 101332T.
Aghaei, F., M. Tan, A. B. Hollingsworth, and B. Zheng. Applying a new quantitative global breast MRI feature analysis scheme to assess tumor response to chemotherapy. J. Magn. Resonance Imaging 44(5):1099–1106, 2016.
Athanassiadi, K., A. Makrygianni, E. Balis, N. Alevizopoulos, M. Vaslamatzis, and C. Vourlakou. False- positive and false-negative rate after positron emission tomography/computer tomography scan for mediastinal staging in non-small-cell lung cancer. Eur. Respir. J. 42(1):93–100, 2014.
Badrinarayanan, V., A. Kendall, and R. Cipolla. SegNet: a deep convolutional encoder-decoder architecture for scene segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39(99):2481–2495, 2015.
Balasubramanian, T., S. Krishnan, M. Mohanakrishnan, K. R. Rao, C. V. Kumar, and K. Nirmala. Hog feature based SVM classification of glaucomatous fundus image with extraction of blood vessels. In: India Conference, 2017, pp. 1–4.
Boris, G., P. Jean Michel, B. Franck, L. Sylvain, G. Sverine, C. Jean-Pierre, K. Denis, H. Patrick, B. Christophe, and C. Bruno. Visceral fat area is an independent predictive biomarker of outcome after first-line bevacizumab-based treatment in metastatic colorectal cancer. Gut 59(3):341–347, 2010.
Brebisson, A. D. and G. Montana. Deep neural networks for anatomical brain segmentation. In: Computer Vision & Pattern Recognition Workshops, vol. 2015-October, 2015, pp. 20–28.
Caprio, S. Relationship between abdominal visceral fat and metabolic risk factors in obese adolescents. Am. J. Hum. Biol. 11(2):259–266, 1999.
Chandra, M. A. and S. S. Bedi. Survey on svm and their application in image classification. Int. J. Inform. Technol. 2:1–11, 2018.
Chen, L. C., G. Papandreou, I. Kokkinos, K. Murphy, and A. L. Yuille. DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFS. IEEE Trans. Pattern Anal. Mach. Intell. 40(4):834–848, 2018.
Chen, L. C., G. Papandreou, F. Schroff, and H. Adam. Rethinking atrous convolution for semantic image segmentation. arXiv:1706.05587, 2017.
Chen, L. C., Y. Zhu, G. Papandreou, F. Schroff, and H. Adam. Encoder–decoder with atrous separable convolution for semantic image segmentation. In: European Conference on Computer Vision, pp. 833–851, 2018.
Commandeur, F., M. Goeller, J. Betancur, S. Cadet, M. Doris, C. Xi, D. S. Berman, P. J. Slomka, B. K. Tamarappoo, and D. Dey. Deep learning for quantification of epicardial and thoracic adipose tissue from non-contrast CT. IEEE Trans. Med. Imaging 37(8):1835–1846, 2018.
Dalal, N. and B. Triggs. Histograms of oriented gradients for human detection. In: IEEE Computer Society Conference on Computer Vision & Pattern Recognition, vol. 2, 2005.
Després, J. P., I. Lemieux, J. Bergeron, P. Pibarot, P. Mathieu, E. Larose, J. RodésCabau, O. F. Bertrand, and P. Poirier. Abdominal obesity and the metabolic syndrome: contribution to global cardiometabolic risk. Arterioscler. Thromb. Vasc. Biol. 28(6):1039, 2008.
Drozdzal, M., E. Vorontsov, G. Chartrand, S. Kadoury, and C. Pal. The importance of skip connections in biomedical image segmentation. arXiv:1608.04117, 2016.
Emaminejad, N., W. Qian, Y. Guan, M. Tan, Y. Qiu, H. Liu, and B. Zheng. Fusion of quantitative image and genomic biomarkers to improve prognosis assessment of early stage lung cancer patients. IEEE Trans. Biomed. Eng. 63(5):1034–1043, 2016.
Estrada, S., R. Lu, S. Conjeti, X. Orozco-Ruiz, J. Panos-Willuhn, M. M. B. Breteler, and M. Reuter. FatSegNet: a fully automated deep learning pipeline for adipose tissue segmentation on abdominal dixon MRI. CoRR. arXiv:abs/1904.02082, 2019.
Fujioka, S., Y. Matsuzawa, K. Tokunaga, and S. Tarui. Contribution of intra-abdominal fat accumulation to the impairment of glucose and lipid metabolism in human obesity. Metab. Clin. Exp. 36(1):54–59, 1987.
Furey, T. S., Cristianini, N., Duffy, N., D. W. Bednarski, M. Schummer, and D. Haussler. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics 16(10):906–914, 2000.
S. Hai, F. Liu, Y. Xie, F. Xing, S. Meyyappan, and Y. Lin. Region segmentation in histopathological breast cancer images using deep convolutional neural network. In: IEEE International Symposium on Biomedical Imaging, 2015, pp. 55–58.
He, Y., M. Keuper, B. Schiele, and M. Fritz. Learning dilation factors for semantic segmentation of street scenes. In: LNCS, vol. 10496, pp. 41–51, 2017.
Hill, J. E., M. Fernandez-Del-Valle, R. Hayden, and S. Mitra. An automated segmentation for direct assessment of adipose tissue distribution from thoracic and abdominal dixon-technique mr images. In: Society of Photo-optical Instrumentation Engineers, vol. 10133, 2017, p. 1013315.
Hinton, G. and T. Tieleman. Lecture 6.5-rmsprop: divide the gradient by a running average of its recent magnitude. COURSERA 4:26–30, 2012.
https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.train_test_split/.
Huang, G., Z. Liu, V. D. M. Laurens, and K. Q. Weinberger. Densely connected convolutional networks. In: European Conference on Computer Vision, vol. 2017-January, pp. 2261–2269, 2016.
Hui, S. C. N., T. Zhang, L. Shi, D. Wang, and W. C. W. Chu. Automated segmentation of abdominal subcutaneous adipose tissue and visceral adipose tissue in obese adolescent in MRI. Mag. Reson. Imaging 45:97–104, 2017.
Ioffe, S. and C. Szegedy. Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv:1502.03167, 2015.
Jegou, S., M. Drozdzal, D. Vazquez, A. Romero, and Y. Bengio. The one hundred layers tiramisu: fully convolutional dense nets for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, pp. 1175–1183, 2016.
Ke, Y., X. Wang, L. Le, R. M. Summers, Y. Ke, X. Wang, L. Le, R. M. Summers, Y. Ke, and X. Wang. Deeplesion: automated deep mining, categorization and detection of significant radiology image findings using large-scale clinical lesion annotations. arXiv:1710.01766, 2017.
Kim, S. H., J. H. Lee, B. Ko, and J. Y. Nam. X-ray image classification using random forests with local binary patterns. In: International Conference on Machine Learning & Cybernetics, vol. 6(July), 2010, pp. 3190–3194.
Kissebah, A. H. and A. N. Peiris. Biology of regional body fat distribution: relationship to non-insulin-dependent diabetes mellitus. Diabetes Metab. Rev. 5(2):83–109, 2010.
Kissebah, A. H., N. Vydelingum, R. Murray, D. J. Evans, A. J. Hartz, R. K. Kalkhoff, and P. W. Adams. Relation of body fat distribution to metabolic complications of obesity. J. Clin. Endocrinol. Metab. 54(2):254–254, 1982.
Klein, S., U. van-der Heide, I. Lips, M. van Vulpen, M. Staring, and J. Pluim. Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information. Med. Phys. 35(4):1407–1417, 2008.
Kullberg, J., A. Hedström, J. Brandberg, R. Strand, L. Johansson, G. Bergström, and H. Ahlström. Automated analysis of liver fat, muscle and adipose tissue distribution from CT suitable for large-scale studies. Sci. Rep. 7(1):10425, 2017.
Kumar, K. V. V. and P. V. V. Kishore. Indian classical dance mudra classification using hog features and svm classifier. Int. J. Electr. Comput. Eng. 7(5):2537, 2018.
Kvist, H., L. Sjoestroem, B. Chowdhury, M. Alpsten, B. Arvidsson, L. Larsson, and A. Cederblad. Determination of total adipose tissue and body fat in women by computed tomography, 40k, and tritium. Am. J. Physiol. 250(6 Pt 1):E736, 1986.
Langer, T., A. Hedstrom, K. Morwald, D. Weghuber, A. Forslund, P. Bergsten, H. Ahlstrom, and J. Kullberg. Fully convolutional networks for automated segmentation of abdominal adipose tissue depots in multicenter water-fat MRI. Magn. Reson. Med. 81(4)2736–2745, 2019.
Larsson, B., K. Svärdsudd, L. Welin, L. Wilhelmsen, P. Björntorp, and G. Tibblin. Abdominal adipose tissue distribution, obesity, and risk of cardiovascular disease and death: 13 year follow up of participants in the study of men born in 1913. Br. Med. J. 288(6428)1401–1404, 1984.
Li, Z. and Y. Yu. Protein secondary structure prediction using cascaded convolutional and recurrent neural networks. In: Conference: International Joint Conference on Artificial Intelligence (IJCAI), New York, 2016.
Liu, J., F. Chen, C. Pan, M. Zhu, X. Zhang, L. Zhang, and H. Liao. A cascaded deep convolutional neural network for joint segmentation and genotype prediction of brainstem gliomas. IEEE Trans. Bio-Med. Eng. 99:1, 2018.
Makrogiannis, S., G. Caturegli, C. Davatzikos, and L. Ferrucci. Computer-aided assessment of regional abdominal fat with food residue removal in CT. Acad. Radiol. 20(11)1413–1421, 2013.
Martinezuseros, J. and J. Garciafoncillas. Obesity and colorectal cancer: molecular features of adipose tissue. J. Transl. Med. 14(1):1–12, 2016.
Mensink, S. D., J. W. Spliethoff, R. Belder, J. M. Klaase, R. Bezooijen, and C. H. Slump. Development of automated quantification of visceral and subcutaneous adipose tissue volumes from abdominal CT scans. In: Medical Imaging 2011: Computer-Aided Diagnosis, vol. 7963, no. 0, p. 79632Q, 2011.
Moses, L. E., D. Shapiro, and B. Littenberg. Combining independent studies of a diagnostic test into a summary ROC curve: data-analytic approaches and some additional considerations. Stat. Med. 12(14):1293–1316, 1993.
Mun, E. C., G. L. Blackburn, and J. B. Matthews. Current status of medical and surgical therapy for obesity. Gastroenterology 120(3):669–681, 2001.
Nakamura, T., K. Tokunaga, I. Shimomura, M. Nishida, S. Yoshida, K. Kotani, A. H. M. W. Islam, Y. Keno, T. Kobatake, and Y. Nagai. Contribution of visceral fat accumulation to the development of coronary artery disease in non-obese men. Atherosclerosis 107(2):239–246, 1994.
Neumann, D., T. Langner, F. Ulbrich, D. Spitta, and D. Goehring. Online vehicle detection using haar-like, LBP and HOG feature based image classifiers with stereo vision preselection. In: Proceedings of the on Intelligent Vehicles Symposium, 2017.
Ochs, R., J. Goldin, F. Abtin, H. Kim, K. Brown, P. Batra, D. Roback, M. Mcnitt-Gray, and M. Brown. Automated classification of lung bronchovascular anatomy in CT using adaboost. Med. Image Anal. 11(3):315–324, 2007.
Ogden, C. L., M. D. Carroll, B. K. Kit, and K. M. Flegal. Prevalence of childhood and adult obesity in the united states, 2011–2012. JAMA 311(8):806, 2014.
Oquab, M., L. Bottou, I. Laptev, and J. Sivic. Learning and transferring mid-level image representations using convolutional neural networks. In: IEEE Conference on Computer Vision & Pattern Recognition, 2014, pp. 1717–1724.
Palacharla, P. K. Machine learning driven model inversion methodology to detect reniform nematodes in cotton. Dissertations & Theses - Gradworks, 2011.
Pedregosa, F., G. Varoquaux, A. Gramfort, V. Michel, and G. Louppe. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12(10):2825–2830, 2013.
Peiris, A. N., M. S. Sothmann, R. G. Hoffmann, M. I. Hennes, C. R. Wilson, A. B. Gustafson, and A. H. Kissebah. Adiposity, fat distribution, and cardiovascular risk. Ann. Int. Med. 110(11):867–872, 1989.
Pomponiu, V., H. Hariharan, B. Zheng, and D. Gur. Improving breast mass detection using histogram of oriented gradients. In: Medical Imaging: Computer-Aided Diagnosis, vol. 9035, 2014, p. 90351R.
Qiu, Y., M. Tan, S. Mcmeekin, T. Thai, K. Ding, K. Moore, H. Liu, and B. Zheng. Early prediction of clinical benefit of treating ovarian cancer using quantitative ct image feature analysis. Acta Radiol. 57(9):1149, 2016.
Rajendran, P. and M. Madheswaran. Hybrid medical image classification using association rule mining with decision tree algorithm. Comput. Sci. 3(10):1173–1178, 2010.
Romero, D., J. C. Ramirez, and A. Marmol. Quantification of subcutaneous and visceral adipose tissue using CT. In: IEEE International Workshop on Medical Measurement & Applications, 2006, pp. 128–133.
Ronneberger, O., P. Fischer, and T. Brox. U-net: convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing & Computer-assisted Intervention, vol. 9351, 2015, pp. 234–241.
Saha, S., A. Mahmud, A. A. Ali, and M. A. Amin. Classifying digital X-ray images into different human body parts. In: International Conference on Informatics, 2016, pp. 67–71.
Shelhamer, E., J. Long, and T. Darrell. Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39(4):640–651, 2014.
Shen, N., X. Li, S. Zheng, L. Zhang, Y. Fu, X. Liu, M. Li, J. Li, S. Guo, and H. Zhang. Automated and accurate quantification of subcutaneous and visceral adipose tissue from magnetic resonance imaging based on machine learning. Magn. Reson. Imaging. https://doi.org/10.1016/j.mri.2019.04.007, 2019.
Sinno Jialin, P. and Y. Qiang. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 22(10):1345–1359, 2010.
Slaughter, K. N., T. Thai, S. Penaroza, D. M. Benbrook, E. Thavathiru, K. Ding, T. Nelson, D. S. Mcmeekin, and K. N. Moore. Measurements of adiposity as clinical biomarkers for first-line bevacizumab-based chemotherapy in epithelial ovarian cancer. Gynecol. Oncol. 133(1):11–15, 2014.
Spasojević, A., O. Stojanov, T. L. Turukalo, and O. Sveljo, Estimation of subcutaneous and visceral fat tissue volume on abdominal MR images, 2015, pp. 217–220.
Suykens, J. A. K. and J. Vandewalle. Least squares support vector machine classifiers. Neural Process. Lett. 9(3):293–300, 1999.
Tajbakhsh, N., S. R. Gurudu, and J. Liang. Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans. Med. Imaging 35(2):630–644, 2016.
Tokunaga, K., Y. Matsuzawa, K. Ishikawa, and S. Tarui. A novel technique for the determination of body fat by computed tomography. Int. J. Obes. 7(5):437–445, 1983.
Van, D. W. S., J. L. Schönberger, J. Nuneziglesias, F. Boulogne, J. D. Warner, N. Yager, E. Gouillart, T. Yu, and T. S. Contributors. Scikit-image: image processing in python. PeerJ 2(2):e453, 2014.
Walid, Z., T. Brown, A. Murtada, and S. Ali. The application of deep learning to quantify SAT/VAT in human abdominal area. In: Advances in Science and Engineering Technology International Conferences (ASET), 2019, pp. 1–5.
Wang, P., X. Hu, Y. Li, Q. Liu, and X. Zhu. Automatic cell nuclei segmentation and classification of breast cancer histopathology images. Signal Process. 122:1–13, 2016.
Wang, Y., Y. Qiu, T. Thai, K. Moore, L. Hong, and B. Zheng. A two-step convolutional neural network based computer-aided detection scheme for automatically segmenting adipose tissue volume depicting on CT images. Comput. Methods Program. Biomed. 144:97–104, 2017.
Wang, Y., T. Thai, K. Moore, K. Ding, S. Mcmeekin, H. Liu, and B. Zheng. Quantitative measurement of adiposity using CT images to predict the benefit of bevacizumab-based chemotherapy in epithelial ovarian cancer patients. Oncol. Lett. 12(1):680–686, 2016.
Wu, X. An iterative convolutional neural network algorithm improves electron microscopy image segmentation. Comput. Sci., pp. 1–9, 2015.
Xie, J., L. Yang, S. C. Zhu, and N. W. Ying. A theory of generative convnet. In: International Conference on International Conference on Machine Learning, 2016.
Yi, L. and Y. F. Zheng. One-against-all multi-class SVM classification using reliability measures. In: IEEE International Joint Conference on Neural Networks, vol. 2, 2013, pp. 849-854.
Yoon, D. Y., J. H. Moon, H. K. Kim, C. S. Choi, S. K. Chang, E. J. Yun, and Y. L. Seo. Comparison of low-dose ct and mr for measurement of intra-abdominal adipose tissue 1: a phantom and human study. Acad. Radiol. 15(1):62–70, 2008.
Yoshizumi, T., T. Nakamura, M. Yamane, A. H. Islam, M. Menju, K. Yamasaki, T. Arai, K. Kotani, T. Funahashi, and S. Yamashita. Abdominal fat: standardized technique for measurement at CT. Radiology 211(1):283–286, 1999.
Yu, L., H. Chen, Q. Dou, J. Qin, and P. A. Heng. Integrating online and offline 3D deep learning for automated polyp detection in colonoscopy videos. IEEE J. Biomed. Health Inform. 21(1):65–75, 2017.
Yu, F. and V. Koltun. Multi-scale context aggregation by dilated convolutions. In: Conference Paper at ICLR, pp. 1–9, 2016.
Zhao, H., J. Shi, X. Qi, X. Wang, and J. Jia. Pyramid scene parsing network. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), vol. 2017-January, 2017, pp. 6230–6239.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (under Grants 61375063, 61271355, 11301549 and 11271378) and also funded by the Graduate Student Innovation Foundation of Central South University (2019zzts213).
Conflict of interest
All authors declared that they have no conflict of interests.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Associate Editor Andreas Anayiotos oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A: Experiments on NIH DeepLesion
Appendix A: Experiments on NIH DeepLesion
We further report segmentation of visceral and subcutaneous adipose tissue results on the DeepLesion dataset.30 This dataset was collected from the PACS of a major medical institute. It contains over 30K radiological images (mostly 512 × 512). To validate our method, 290 abdominal CT images are randomly selected to split into three subsets: training subset (total of 203 images), including a validation subset, and testing subset (total of 87 images). All images are stored in unsigned 16 bit, and manually label by radiologists from the Second XiangYa Hospital of Central South University.
All of the abdominal CT images are standardized to 512 × 512. Then, apply our proposed framework without modifications, including the computer conditions and parameters of experiments. Total of 12 images from testing subset are randomly selected to display the performance of our proposed framework. Figure 10 shows the results with red color in VAT and green color in SAT.
Rights and permissions
About this article
Cite this article
Wang, Z., Meng, Y., Weng, F. et al. An Effective CNN Method for Fully Automated Segmenting Subcutaneous and Visceral Adipose Tissue on CT Scans. Ann Biomed Eng 48, 312–328 (2020). https://doi.org/10.1007/s10439-019-02349-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-019-02349-3