Abstract
Syntrophic interspecies electron exchange is essential for the stable functioning of diverse anaerobic microbial communities. Hydrogen/formate interspecies electron transfer (HFIT), in which H2 and/or formate function as diffusible electron carriers, has been considered to be the primary mechanism for electron transfer because most common syntrophs were thought to lack biochemical components, such as electrically conductive pili (e-pili), necessary for direct interspecies electron transfer (DIET). Here we report that Syntrophus aciditrophicus, one of the most intensively studied microbial models for HFIT, produces e-pili and can grow via DIET. Heterologous expression of the putative S. aciditrophicus type IV pilin gene in Geobacter sulfurreducens yielded conductive pili of the same diameter (4 nm) and conductance of the native S. aciditrophicus pili and enabled long-range electron transport in G. sulfurreducens. S. aciditrophicus lacked abundant c-type cytochromes often associated with DIET. Pilin genes likely to yield e-pili were found in other genera of hydrogen/formate-producing syntrophs. The finding that DIET is a likely option for diverse syntrophs that are abundant in many anaerobic environments necessitates a reexamination of the paradigm that HFIT is the predominant mechanism for syntrophic electron exchange within anaerobic microbial communities of biogeochemical and practical significance.
Similar content being viewed by others
Introduction
One of the most significant unanswered questions in the microbial ecology of important methanogenic environments such as anaerobic soils/sediments and anaerobic digesters is the relative importance of direct interspecies electron transfer (DIET) versus H2/formate interspecies electron transfer (HFIT). DIET and HFIT are strategies for the syntrophic anaerobic oxidation of key intermediates (organic acids, alcohols, and aromatics) in the conversion of organic matter to methane [1,2,3,4,5]. The relative proportion of electron flux through DIET or HFIT can influence the speed of interspecies electron transfer, the stability of anaerobic microbial communities, and their ability to adapt to environmental change [3, 4, 6, 7]. However, there are no accurate methods for measuring the rates at which H2 and formate are transferred between microbes [8] or for quantifying in situ interspecies electrical currents in complex communities.
Interspecies H2 [9] and formate [8, 10] transfer (see [11] for comprehensive review) were conceived long before DIET [12, 13]. Therefore, for decades, interactions within complex anaerobic microbial communities were interpreted through the lens of HFIT. However, reexamination of key environmental data suggests that the results were more generally consistent with DIET. For example, estimated H2 turnover rates in anaerobic digesters, rice paddy soils, and aquatic sediments were consistently less than 10% of the independently determined rate of methane production derived from the reduction of carbon dioxide to methane [14,15,16]. This result is consistent with DIET providing most of the electrons for carbon dioxide reduction. However, DIET was not yet a concept and the mismatch between measured H2 turnover rates, and the assumed role of H2 as a primary interspecies electron carrier, was rationalized with the suggestion that there was a pool of H2 within closely juxtapositioned assemblages of H2 producers and H2 consumers with higher H2 concentrations than the H2 pool in the bulk aqueous environment. The existence of two distinct pools of H2 that did not equilibrate over time via diffusion, a seemingly physical impossibility, was never verified.
An evaluation of potential methods for detecting DIET concluded that there is yet not sufficient information on which microorganisms are capable of DIET to determine the importance of DIET based on the composition of microbial communities and their gene or protein expression patterns [5]. Geobacter species are the only bacteria demonstrated to function as the electron-donating partners for DIET [13, 17,18,19,20,21,22] and are abundant and metabolically active in environments in which DIET is likely [19, 23,24,25]. However, in many instances in which DIET is thought to be important other bacteria, not closely related to Geobacter species, predominate [2,3,4,5,6,7]. Experimental evaluation of microbial isolates for their ability to participate in DIET is slow and laborious because defined co-cultures established with isolates that have not been routinely grown via DIET can take a long time to adapt to DIET. Pre-selection of likely candidates with screening methods that identify cell characteristics that may be associated with DIET could lead to more positive results in DIET co-culture tests.
Extracellular electron transport proteins thought to be important for DIET in Geobacter include electrically conductive pili (e-pili) and outer-surface c-type cytochromes [13, 18, 19, 21, 26]. The presence of outer-surface c-type cytochromes is a poor indicator for the ability to participate in DIET. Several Geobacter species, with genes for outer-surface c-type cytochromes did not grow via DIET in defined co-cultures [20] and Shewanella oneidensis has not been grown via DIET despite its outer-surface c-type cytochromes that enable other forms of extracellular electron transfer [27].
The ability of microbes to express e-pili is possibly a better predictor of the potential to participate in DIET. Geobacter metallireducens expressing its native e-pili or the e-pili of G. sulfurreducens served as the electron-donating partner for DIET, but a G. metallireducens strain expressing poorly conductive pili could not [26]. G. uraniireducens, which expresses genes for outer-surface c-type cytochromes when grown with insoluble electron acceptors [28, 29], could not be grown via DIET [20], presumably because its pili are poorly conductive [30, 31].
It was initially considered that only short pilin monomers (ca. 60 amino acids), such as those found in G. metallireducens and G. sulfurreducens, could assemble into e-pili [12, 32]. However, larger pilin monomers (>100 amino acids), phylogenetically distinct from Geobacter pilins, can yield e-pili and e-pili have independently evolved multiple times [33]. This raises the possibility that some microorganisms not previously known to be capable of extracellular electron transfer may express e-pili to enable DIET.
Close packing of aromatic amino acids appears to be important for electron transport along G. sulfurreducens e-pili [31, 34,35,36,37,38,39], the e-pili of G. metallireducens [40], the conductive archaellum of Methanospirillum hungatei [41], and pili assembled from synthetic pilin genes [42, 43]. From this preliminary understanding of e-pili conductivity mechanisms and conductivity measurements on pili expressed from a diversity of pilins, empirical criteria to predict whether a pilin is likely to be assembled into e-pili were established [33]. The criteria are: (1) aromatic amino acids are located in the key positions required for conductivity in G. sulfurreducens e-pili; (2) the abundance of aromatic amino acids is above the minimum threshold of 9% found in e-pili described to date; and (3) no large gaps (>40 amino acids) within the pilin that lack aromatic amino acids.
Syntrophus aciditrophicus is one of the most intensively studied pure culture models for HFIT [44,45,46,47]. It was previously concluded that S. aciditrophicus would be unlikely to participate in DIET because it lacks a gene encoding a protein homologous to the Geobacter pilin monomer [46, 48]. Here we report that S. aciditrophicus expresses e-pili and is capable of growing via DIET. These results, and analysis of the pilin genes of other common syntrophs, indicate that the capacity for DIET should be considered as an option for microorganisms known to grow via HFIT, suggesting that DIET may be more widespread than previously considered.
Results and discussion
Transmission electron microscopy of S. aciditrophicus revealed filaments with a morphology typical of type IVa pili (Fig. 1a). A complement of genes for protein components required for type IV pilus assembly (pilA, pilD, pilM, pilN, pilO, pilP, pilQ) and transcriptional control of pilus expression (pilS, pilR) are also present in the genome (Supplementary Fig. 1). One gene, SYN 00814 encodes a peptide with a N-terminal domain characteristic of PilA, the pilin monomer for Type IVa pili found in other microorganisms (Fig. 1b). This includes a short leader peptide (13 amino acids) that is cleaved by PilD at the G|FTLIE recognition site and a highly conserved, hydrophobic, pilin assembly interface that facilitates pilin polymerization into pili [49, 50].
a Transmission electron micrograph showing multiple pili protruding from S. aciditrophicus (pili highlighted with black arrows). b Key characteristics of the predicted amino acid sequence of the S. aciditrophicus pilin monomer. The PilA monomer of G. sulfurreducens that yields conductive pili is shown for comparison. Aromatic amino acid abundance is calculated as a percentage of the mature peptide.
The amino acid sequence of the putative PilA fits the empirical criteria [33] for a pilin monomer likely to yield e-pili, listed in the Introduction. The first five aromatic amino acids of the S. aciditrophicus pilin are in the same position as the first five aromatic amino acids of the G. sulfurreducens pilin (Fig. 1b). The abundance of aromatic amino acids (10.9% of amino acids) is above the minimum threshold of 9% thought to be necessary for high e-pili conductivity. The largest gap between aromatic amino acids in the pilin sequence is 22 amino acids, well below the largest aromatic-free gap (<40 amino acids) found to be allowable for the formation of e-pili in previous studies [33].
Low culture densities of S. aciditrophicus prevented harvesting sufficient quantities of pili to measure pili conductance on the electrode arrays previously employed for the study of other e-pili [33]. Therefore, the method initially employed to document the presence of e-pili in G. sulfurreducens [12] was adapted [41] as an alternative approach. Cultures of S. aciditrophicus were directly drop cast on highly ordered pyrolytic graphite (HOPG), washed with deionized water, and examined with atomic force microscopy. Topographic imaging, (Fig. 2a), indicated that the distance from the surface of the HOPG to the top of the pili (Fig. 2b) was 4.0 ± 0.7 nm (n = 27; nine different locations on three separate pili). The 4 nm diameter of the S. aciditrophicus pili is larger than the 3 nm reported for the e-pili of Geobacter species [12, 34, 36, 40, 42], but thinner than the ca. 5–6-nm diameter of the intensively studied type IVa pili of Pseudomonas and Neisseria species [51,52,53].
a Contact mode topographic imaging of pili on highly ordered pyrolytic (HOPG). Red line designates the cross-section examined in b. b Topographic analysis of the height/diameter of an individual pilus and corresponding current measurements (100 mV differential between the tip and the HOPG) across the pilus cross-section. c Current response of the pili shown in a with an applied 100 mV differential. d Current–voltage analysis of individual pili of S. aciditrophicus (blue data points), wild-type G. sulfurreducens (black data points), and the Aro-5 strain of G. sulfurreducens (green data points). Current–voltage spectroscopy is shown for one pilus of each type and is representative of analysis of three distinct locations on three separate pili of each type. Additional scans of S. aciditrophicus pili available in Supplementary Fig. 2. The G. sulfurreducens wild-type and strain Aro-5 data are from reference [41].
Conductive imaging revealed that the pili were electrically conductive (Fig. 2b, c). Point-mode current–voltage (I–V) spectroscopy (Fig. 2d; Supplementary Fig. 2) yielded an Ohmic-like response with a conductance of 6.1 ± 1.2 nS (mean ± standard deviation; n = 9) that was similar to the previously reported [41] conductance of 4.5 ± 0.3 nS for wild-type G. sulfurreducens pili. As previously reported [41], the pili from G. sulfurreducens strain Aro-5, which lack key aromatic amino acids required for high conductivity [34, 36, 37, 39], had much lower (0.004 ± 0.002 nS) conductance (Fig. 2d).
The lack of tools for genetic manipulation of S. aciditrophicus limited further functional analysis of the putative PilA gene predicted to yield its e-pili in the native organism. Therefore, the gene was heterologously expressed in G. sulfurreducens, replacing the native G. sulfurreducens pilA with an approach that has successfully yielded G. sulfurreducens strains that express a diversity of both highly conductive and poorly conductive heterologous pili [31, 33, 40, 43, 54]. This new G. sulfurreducens strain, designated strain SP (for Syntrophus pili), expressed abundant pili (Fig. 3a). Pili preparations sheared from cells and purified, as described previously [33], did not yield a pilin monomer on protein gels under standard protein denaturation conditions with boiling sodium dodecyl sulfate (Supplementary Fig. 4), but did yield a single band of the molecular weight (13 kDa) expected for the S. aciditrophicus pilin monomer on denaturing gels when the pili were first treated with a strong detergent (Supplementary Fig. 4) known to be required to depolymerize G. sulfurreducens e-pili [55]. When this band was excised and analyzed with mass spectrometry the only peptides recovered were four peptides that are exclusively unique to the S. aciditrophicus pilin (Supplementary Fig. 4).
a Transmission electron micrograph of pili expression in the strain of G. sulfurreducens in which the native pilA was replaced with the S. aciditrophicus pilA. Examples of the location of pili are designated with white arrows. b Current–voltage profile for three separate locations on three pili expressed by S. aciditrophicus pili (data aggregated from individual plots in Fig.1d and Supplementary Fig. 2) and for three separate locations on three pili expressed in the SP strain of G. sulfurreducens in which the wild-type pilA was replaced with the S. aciditrophicus pilA (individual plots shown in Supplementary Fig. 3). c Current production by G. sulfurreducens with wild-type PilA; the synthetic Aro-5 PilA designed to yield poorly conductive pili; or S. aciditrophicus PilA. d Four-probe conductance (mean + standard deviation n = 9) of films of purified pili from strains of G. sulfurreducens expressing; S. aciditrophicus PilA compared with previously reported [33] conductances of pili derived from wild-type and Aro-5 strains of G. sulfurreducens.
Also consistent with the heterologous expression of S. aciditrophicus pili was the finding that the pili emanating from G. sulfurreducens strain SP had a diameter (4.1 ± 0.4 nm) and conductance (5.9 ± 0.7 nS) nearly identical to the native pili expressed by S. aciditrophicus (Fig. 3b). Although preparations of outer-surface proteins from G. sulfurreducens contain filaments comprised of OmcS that also have a 4 nm diameter [56], these OmcS filaments may be an artifact of the method employed for harvesting the proteins because only filaments of 3 nm are observed when cells are directly drop cast on HOPG without any pretreatment [30]. We reverified this by measuring filaments the heights of pili from wild-type G. sulfurreducens drop cast on HOPG. The heights were 3 + 0.1 (n = 18, six different locations on three separate pili).
Furthermore, G. sulfurreducens strain SP produced electrical current densities comparable with the control strain expressing the G. sulfurreducens wild-type pilA (Fig. 3c). Such high current densities are only possible when G. sulfurreducens expresses e-pili [33, 39]. As previously reported [33, 39], G. sulfurreducens strain Aro-5, with its poorly conductive pili, produced much lower current densities (Fig. 3c). Networks of pili sheared from the electrode-grown biofilm of strain SP, purified, and drop cast on electrode arrays as previously described [33], had a conductance comparable with G. sulfurreducens wild-type pili networks (Fig. 3d). These results further demonstrated that the S. aciditrophicus PilA gene yields a pilin monomer that can assemble into e-pili.
The presence of e-pili in S. aciditrophicus suggested it might be capable of establishing an electrical connection for DIET. To evaluate this, S. aciditrophicus was grown in co-culture with G. sulfurreducens, the microbe most intensively studied as an electron-accepting partner for DIET [3]. G. sulfurreducens can also function as a H2- and formate-consuming partner for HFIT [57]. Therefore, even if S. aciditrophicus was unable to grow via DIET it would be expected to grow in co-culture with G. sulfurreducens via HFIT, providing a positive control to ensure that the culture medium was appropriate for co-culture growth. The electron donor was benzoate, a substrate that S. aciditrophicus can metabolize, but G. sulfurreducens cannot. The electron acceptor was fumarate, an electron acceptor that only G. sulfurreducens can utilize. In the presence of a H2/formate-consuming partner, S. aciditrophicus metabolizes benzoate to acetate with the production of either H2:
or formate:
With electron transfer via DIET the relevant reaction is:
In addition to oxidizing H2 and formate, or consuming electrons released during DIET, G. sulfurreducens can also metabolize acetate, making the overall reaction expected for the oxidation of benzoate with the reduction of fumarate to succinate in the co-culture:
S. aciditrophicus/G. sulfurreducens co-cultures grew with repeated sub-culturing and, within the experimental error, exhibited the expected stoichiometry of benzoate consumption and succinate production (Fig. 4a). In these co-cultures HFIT, DIET, or a combination of the two, were feasible. Therefore, to eliminate the possibility of HFIT, S. aciditrophicus was co-cultured with the previously described strain of G. sulfurreducens [21], designated here as G. sulfurreducensHF, that cannot utilize H2 or formate because the genes for the uptake hydrogenase and formate dehydrogenase were deleted. The S. aciditrophicus/G. sulfurreducensHF co-culture had a longer initial lag period, but then metabolized benzoate with the reduction of fumarate almost as fast as the co-culture with wild-type G. sulfurreducens (Fig. 4b). These results demonstrate that S. aciditrophicus can grow via DIET.
Metabolism with S. aciditrophicus co-cultured with (a) wild-type G. sulfurreducens, (b) G. sulfurreducensHF, which is unable to use H2 or formate. c G. sulfurreducensHF with granular activated carbon (GAC) amendment. No acetate or formate was detected in any of the co-cultures. GAC interfered with determination of benzoate, which is not shown for GAC-amended cultures. Scanning electron micrographs (SEM) of cells collected on filters from co-cultures with (d) G. sulfurreducens wild-type or (e) G. sulfurreducens strainHF demonstrating greater aggregation in co-cultures with G. sulfurreducensHF in which DIET was the only option for interspecies electron exchange. f SEM of cells on GAC from co-culture of S. aciditrophicus with G. sulfurreducens strainHF. Circles-benzoate; squares-succinate. Data are the mean and standard deviation of triplicate cultures.
DIET requires physical electrical contacts between the electron-donating and electron-accepting partner, whereas contact is not required for HFIT [2, 3]. In some co-cultures the requirement for contact manifests as visible aggregates [13], but other DIET co-cultures produce small, relatively fragile aggregates [18]. There were no visible aggregates in the S. aciditrophicus/G. sulfurreducens co-cultures. However, more small clumps were seen in scanning electron micrographs of co-cultures in which G. sulfurreducensHF was the electron-accepting partner than co-cultures with wild-type G. sulfurreducens provided as the electron-accepting partner (Fig. 4 d, e). This observation is consistent with the need for contact between electron-donating and electron-accepting partners participating in DIET. In contrast, HFIT was an option for the co-cultures established with wild-type G. sulfurreducens and aggregation is not required when G. sulfurreducens grows via HFIT [57].
Granular activated carbon (GAC) can greatly reduce the initial lag time in establishing DIET-based co-cultures because both partners attach to GAC, which functions as an electrically conductive conduit [18, 58]. GAC substantially reduced the lag time of the S. aciditrophicus/G. sulfurreducensHF co-cultures (Fig. 4c). As previously observed for other co-cultures in which GAC promoted DIET, cells from the S. aciditrophicus/G. sulfurreducensHF co-culture heavily colonized the GAC (Fig. 4e), consistent with a GAC conduit for DIET.
In addition to e-pili, multi-heme outer-surface c-type cytochromes appear to play an important role in DIET in Geobacter species [2, 3]. However, the S. aciditrophicus genome encodes only a few putative c-type cytochromes [46] and cytochromes were not readily apparent in heme-stained preparations of cell protein (Supplementary Fig. 5). Not all microbes require cytochromes for effective electron transport to the outer cell surface [59]. More detailed examination of the role of e-pili and other S. aciditrophicus components in DIET will require the development of methods for genetic manipulation of this microorganism.
Implications
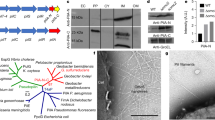
S. aciditrophicus is the first isolate outside the genus Geobacter definitely demonstrated to function as the electron-donating partner for DIET and it is the first syntroph shown to have the option to grow via HFIT or DIET. Other Syntrophus species also have pilin genes with the empirical criteria, detailed in the “Introduction”, of aromatic amino acid placement and abundance likely to yield e-pili (Fig. 5). The pilins of other diverse genera of syntrophic microorganisms known to grow via HFIT in defined co-cultures also meet these criteria (Fig. 5). Establishing conditions that favor DIET often enrich for microbes in these genera [4]. For example, in enrichment cultures specifically designed to promote the metabolism of propionate or butyrate via DIET, Smithella (propionate enrichment) or Syntrophomonas (butyrate enrichment) species were the most abundant bacteria [60,61,62]. The greater energetic demands [63] required for synthesizing the abundant aromatic amino acids that are required for e-pili conductivity suggests that e-pili provide a strong selective advantage under some environmental conditions. Conferring the capacity for DIET is a likely explanation.
Position of aromatic amino acids designated in magenta and the largest gap that lacks aromatic amino acids designated in blue. The full protein sequence alignment for each pilin can be found in Supplementary Fig. 6. Each of the pilins meets the empirical criteria derived from previous studies [33] of: (1) aromatic amino acids located in the key positions required for conductivity in G. sulfurreducens e-pili; (2) the abundance of aromatic amino acids greater than 9%; and (3) no aromatic-free gaps of greater than 40 amino acids.
A fresh perspective and new analytical tools will be required to resolve the relative importance of HFIT and DIET in diverse anaerobic microbial communities. Just as electron-accepting partners have different gene expression patterns depending on whether they are participating in HFIT or DIET [21, 64], it may be possible to determine whether electron-donating syntrophs are engaged in DIET or HFIT from metatranscriptomic analysis of anaerobic communities. However, this approach will require much more information on the diversity of microbes capable of functioning as electron-donating partners for DIET. The recovery of Prosthecochloris aestuarii [17] which can function as an electron-accepting partner for DIET, with an initial enrichment strategys that favored the growth of microorganisms likely to participate in DIET suggests that similar design of appropriate enrichment/isolation approaches may further expand the known diversity of microorganisms that can function as electron-donating partners for DIET.
Methods
Bacterial strains, plasmids, and culture conditions
S. aciditrophicus, G. sulfurreducens wild-type, G. sulfurreducens strain Aro-5, and G. sulfurreducensHF were obtained from our laboratory culture collections. A strain of G. sulfurreducens expressing pilA from S. aciditrophicus rather than native G. sulfurreducens pilA was constructed as previously described [33]. S. aciditrophicus and G. sulfurreducens strains were routinely grown under strict anaerobic conditions at 30 °C in previously described [65] defined, bicarbonate-buffered medium with N2:CO2 (80:20) as the gas phase. For S. aciditrophicus, the medium was amended with crotonate (20 mM) and for G. sulfurreducens strains, acetate (10 mM) was the electron donor and fumarate (40 mM) was the electron acceptor. The presence of c-type cytochromes in whole cell lysates was evaluated with heme staining of proteins separated on denaturing polyacrylamide gels as previously described [26]. G. sulfurreducens strains were grown with a graphite electrode as the electron acceptor as previously described [66].
Co-cultures were established in 10 ml of culture medium [65] in anaerobic pressure tubes with benzoate (1 mM) as the electron donor and fumarate (40 mM) as the electron acceptor with cysteine (2 mM) and sulfide (1 mM) added as reducing agents. When noted, cultures were amended with GAC (0.25 g; 8–20 mesh). Benzoate, acetate, formate, and succinate were analyzed with high-performance liquid chromatography as previously described [22]. Succinate values were corrected for small amounts of succinate produced in controls without benzoate. Previously described methods were employed for transmission electron microscopy [33] and scanning electron microscopy [58].
Characterization of Pili
For analysis of the conductance of individual pili, a 100 µl sample of culture was drop cast onto HOPG. After 10 min, the HOPG was washed twice with 100 µl of deionized water, blotted dry to remove excess water, and allowed to fix for 12 h at 24 °C in a desiccator. Samples were equilibrated with atmospheric humidity for at least 2 h and then examined with an Oxford Instruments Cypher ES Environmental AFM in ORCA electrical mode equipped with a Pt/Ir-coated Arrow-ContPT tip with a 0.2 N/m force constant (NanoWorld AG, Neuchâtel, Switzerland). Pili were located in contact mode, with a set point of 0.002 V and a scan rate of 1.5 Hz. For conductive imaging, the grounded tip, attached to a transimpedance amplifier, served as a translatable top electrode to locally detect the current response of the individual pili to a 100 mV bias applied to the HOPG substrate. Individual pili conductivity was further characterized by lightly pressing the AFM tip (set point 0.002 V) to the top of the pili and applying a quadruplicate amplitude of ±0.6 V voltage sweep at a frequency of 0.99 Hz, receiving ca. 8000 points of reference per measurement. Three independent points (technical replicates) on three individual pili (biological replicates) were used to calculate conductance. Conductance was calculated by using the linear slope between −0.2 and 0.2 V.
Pili expressed in G. sulfurreducens were sheared from cells and further purified as previously described [33]. The conductivity of films of the pili was analyzed with four-probe conductivity measurements on films of pili purified from cells as previously described [33]. Three voltage sweeps were conducted on each device (technical replicates), and three independent devices were tested (biological replicates).
To further confirm the composition of the pili recovered from G. sulfurreducens strain SP, the strain heterologously expressing the pilin gene of S. aciditrophicus, the pili were depolymerized into monomers, with octyl β-D-glucopyranoside (OG), in a modification of the method previously shown to depolymerize G. sulfurreducens e-pili [55]. OG was added to 20 µl of pili suspension to provide a final concentration of 10% OG. After depolymerization for 16 h at room temperature 30 µl of water was added. An equal amount of water was added to 20 µl of pili suspension that was not treated with OG and served as a non-depolymerized control. Both the depolymerized and control samples were mixed with 2 × Laemmli loading buffer (2.1% sodium dodecyl sulfate) containing 5% β-mercaptoethanol, yielding a final OG concentration of 2% in the depolymerized sample. The samples were heated to 95 °C for 10 min and 60 µl of the samples was loaded on to a denaturing gel with 12% SDS and run at 150 V for 60 min on a BioRad mini gel system. The pili monomers were visualized with Coomassie staining. The monomer of expected size was excised and sent for LC MS/MS analysis at the UMass Medical School Mass Spectroscopy Facility.
References
Kouzuma A, Kato S, Wantanabe K. Microbial interspecies interactions: recent findings in syntrophic consortia. Front Microbiol. 2015;6:477.
Lovley DR. Happy together: microbial communities that hook up to swap electrons. ISME J. 2017a;11:327–36.
Lovley DR. Syntrophy goes electric: direct interspecies electron transfer. Ann Rev Microbiol. 2017b;71:643–64.
Martins G, Salvador AF, Pereira L, Alves MM. Methane production and conductive materials: a critical review. Environ Sci Technol. 2018;52:10241–53.
Van Steendam C, Smets I, Skerlos S, Raksin L. Improving anaerobic digestion via direct interspecies electron transfer requires development of suitable characterization methods. Curr Opin Biotechnol. 2019;57:183–90.
Park J-H, Kang H-J, Park K-H, Park H-D. Direct interspecies electron transfer via conductive materials: a perspective for anaerobic digestion applications. Bioresour Technol. 2018;254:300–11.
Barua S, Dhar BR. Advances towards understanding and engineering direct interspecies electron transfer in anaerobic digestion. Bioresour Technol. 2017;244:698–707.
Boone DR, Johnson RL, Liu Y. Diffusion of the interspecies electron carriers H2 and formate in methanogenic ecosystems and its implications in the measurement of Km for H2 or formate uptake. Appl Environ Microbiol. 1989;556:1735–41.
Bryant MP, Wolin EA, Wolin MJ, Wolfe RS. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch Microbiol. 1967;59:20–31.
Thiele JH, Zeikus JG. Control of interspecies electron flow during anaerobic digestion: significance of formate transfer versus hydrogen transfer during syntrophic methanogenesis in flocs. Appl Environ Microbiol. 1988;54:20–29.
Schink B, Stams AJM. Syntrophism among prokaryotes. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes—prokaryotic communities and ecophysiology. Berlin, Germany: Spri9nger-Verlag; 2013. p. 471–93.
Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, Lovley DR. Extracellular electron transfer via microbial nanowires. Nature. 2005;435:1098–101.
Summers ZM, Fogarty H, Leang C, Franks AE, Malvankar NS, Lovley DR. Direct exchange of electrons within aggregates of an evolved syntrophic co-culture of anaerobic bacteria. Science. 2010;330:1413–5.
Conrad R, Babbel M. Effect of dilution on methanogenesis, hydrogen turnover and interspecies hydrogen transfer in anoxic paddy soil. FEMS. 1989;62:21–28.
Conrad R, Mayer H-P, Wust M. Temporal change of gas metabolism by hydrogen-syntrophic methanogenic bacterial associations in anoxic paddy soil. FEMS Microbiol Lett. 1989;62:265–73.
Conrad R, Phelps TJ, Zeikus JG. Gas metabolism evidence in support of juxtaposition of hydrogen-producing and methanogenic bacteria in sewage sludge and lake sediments. Appl Environ Microbiol. 1985;50:595–601.
Ha PT, Lindemann SR, Shi L, Dohnalkova AC, Fredrickson JK, Madigan MT, et al. Syntrophic anaerobic photosynthesis via direct interspecies electron transfer. Nat Commun. 2017;7:13924.
Rotaru A-E, Shrestha PM, Liu F, Nevin KP, Lovley DR. Direct interspecies electron transfer during syntrophic growth of Geobacter metallireducens and Methanosarcina barkeri on ethanol. Appl Environ Microbiol. 2014a;80:4599–605.
Rotaru A-E, Shrestha PM, Liu F, Shrestha M, Shrestha D, Embree M, et al. A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ Sci. 2014b;7:408–15.
Rotaru A-E, Woodard TL, Nevin KP, Lovley DR. Link between capacity for current production and syntrophic growth in Geobacter species. Front Microbiol. 2015;6:744.
Shrestha PM, Rotaru A-E, Summers ZM, Shrestha M, Liu F, Lovley DR. Transcriptomic and genetic analysis of direct interspecies electron transfer. Appl Environ Microbiol. 2013;79:2397–404.
Wang L-Y, Nevin KP, Woodard TL, Mu B-Z, Lovley DR. Expanding the diet for DIET: electron donors supporting direct interspecies electron transfer (DIET) in defined co-cultures. Front Microbiol. 2016;7:236.
Holmes DE, Shrestha PM, Walker DJF, Dang Y, Nevin KP, Woodard TL, et al. Metatranscriptomic evidence for direct interspecies electron transfer between Geobacter and Methanothrix species in rice paddy soils. Appl Environ Microbiol. 2017;83:e00223–17.
Morita M, Malvankar NS, Franks AE, Summers ZM, Giloteaux L, Rotaru AE, et al. Potential for direct interspecies electron transfer in methanogenic wastewater digester aggregates. mBio. 2011;2:e00159–11.
Shrestha PM, Malvankar NS, Werner JJ, Franks AE, Rotaru A-E, Shrestha M, et al. Correlation between microbial community and granule conductivity in anaerobic bioreactors for brewery wastewater treatment. Bioresour Tech. 2014;174:306–10.
Ueki T, Nevin KP, Rotaru A-E, Wang L-Y, Ward JE, Woodard TL, et al. Geobacter strains expressing poorly conductive pili reveal constraints on direct interspecies electron transfer mechanisms. mBio. 2018;9:e01273–18.
Shi L, Dong H, Reguera G, Beyenal H, Lu A, Liu J, et al. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat Rev Microbiol. 2016;14:651–62.
Holmes DE, O'Neil RA, Chavan MA, N'Guessan LA, Vrionis HA, Perpetua LA, et al. Transcriptome analysis of Geobacter uraniireducens growing in uranium-contaminated subsurface sediments. ISME J. 2009;3:216–30.
Aklujkar M, Coppi MV, Leang C, Kim BC, Chavan MA, Perpetua LA, et al. Proteins involved in electron transfer to Fe(III) and Mn(IV) oxides by Geobacter sulfurreducens and Geobacter uraniireducens. Microbiology. 2013;159:515–35.
Lovley DR, Walker DJF. Geobacter protein nanowires. Front Microbiol. 2019;10:2078.
Tan Y, Adhikari RY, Malvankar NS, Ward JE, Nevin KP, Woodard TL, et al. The low conductivity of Geobacter uraniireducens pili suggests a diversity of extracellular electron transfer mechanisms in the genus Geobacter. Front Microbiol. 2016b;7:980.
Holmes DE, Dang Y, Walker DJF, Lovley DR (2016). The electrically conductive pili of Geobacter species are a recently evolved feature for extracellular electron transfer. Microb Genom 2. https://doi.org/10.1099/mgen.1090.000072.
Walker DJF, Adhikari RY, Holmes DE, Ward JE, Woodard TL, Nevin KP, et al. Electrically conductive pili from genes of phylogenetically diverse microorganisms. ISME J. 2018;12:48–58.
Adhikari RY, Malvankar NS, Tuominen MT, Lovley DR. Conductivity of individual Geobacter pili. RSC Adv. 2016;6:8354–7.
Lampa-Pastirk S, Veazey JP, Walsh KA, Feliciano GT, Steidl RJ, Tessmer S, et al. Thermally activated charge transport in microbial protein nanowires. Sci Rep. 2016;6:23517.
Malvankar NS, Yalcin SE, Tuominen MT, Lovley DR. Visualization of charge propagation along individual pili proteins using ambient electrostatic force microscopy. Nat Nanotechnol. 2014;9:1012–7.
Malvankar NS, Vargas M, Nevin KP, Tremblay P-L, Evans-Lutterodt K, Nykypanchuk D, et al. Structural basis for metallic-like conductivity in microbial nanowires. mBio. 2015;6:e00084–15.
Steidl RJ, Lampa-Pastirk S, Reguera G. Mechanistic stratification in electroactive biofilms of Geobacter sulfurreducens mediated by pilus nanowires. Nat Commun. 2016;7:12217.
Vargas M, Malvankar NS, Tremblay P-L, Leang C, Smith JA, Patel P, et al. Aromatic amino acids required for pili conductivity and long-range extracellular electron transport in Geobacter sulfurreducens. mBio. 2013;4:e00105–13.
Tan Y, Adhikari RY, Malvankar NS, Ward JE, Woodard TL, Nevin KP, et al. Expressing the Geobacter metallireducens PilA in Geobacter sulfurreducens yields pili with exceptional conductivity. mBio. 2017;8:e02203–16.
Walker DJF, Martz E, Holmes DE, Zhou Z, Nonnenmann SS, Lovley DR. The archaellum of Methanospirillum hungatei is electrically conductive. mBio. 2019;10:e00579–00519.
Liu X, Wang S, Xu A, Zhang L, Liu H, Ma LZ. Biological synthesis of high-conductive pili in aerobic bacterium Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2019;103:1535–44.
Tan Y, Adhikari RY, Malvankar NS, Pi S, Ward JE, Woodard TL, et al. Synthetic biological protein nanowires with high conductivity. Small. 2016a;12:4481–5.
Hopkins BT, McInerney MJ, Warikoo V. Anaerobic syntrophic benzoate degradation: evidence for a threshold and isolation of a syntrophic benzoate degrader. Appl Environ Microbiol. 1995;61:526–30.
McInerney MJ, Rohlin L, Mouttaki H, Kim U, Krupp RS, Rios-Hernandez L, et al. The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci USA. 2007;104:7600–5.
Sieber JR, Le HM, McInerney MJ. The importance of hydrogen and formate transfer for syntrophic fatty, aromatic and alicyclic metabolism. Environ Microbiol. 2014;16:177–88.
Bradley EJ, Bhupathiraju VK, Tanner RS, Woese CR, McInerney MJ. Syntrophus aciditrophicus sp. nov., a new anaerobic bacterium that degrades fatty acids and benzoate in syntrophic association with hydrogen-using microorganisms. Arch Microbiol. 1999;171:107–14.
Sieber JR, McInerney MJ, Gunsalus RP. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Ann Rev Microbiol. 2012;66:429–52.
Craig L, Li J. Type IV pili: paradoxes in form and function. Curr Opin Struct Biol. 2008;18:267–77.
Hospenthal MK, Costa TRD, Waksman G. A comprehensive guide to pilus biogenesis in Gram-negative bacteria. Nat Rev Microbiol. 2017;15:365–79.
Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, et al. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell. 2006;23:651–62.
Kolappan S, Coureuil M, Yu X, Nassif X, Egelman EH, Craig L. Structure of the Neisseria meningitidis Type IV pilus. Nat Commun. 2016;7:13015.
Wang F, Coureuil M, Osinski T, Orlova A, Altindal T, Gesbert G, et al. Cryoelectron microscopy reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae type IV pili at sub-nanometer resolution. Structure. 2017;25:14231435.
Liu X, Tremblay P-L, Malvankar NS, Nevin KP, Lovley DR, Vargas M. A Geobacter sulfurreducens strain expressing Pseudomonas aeruginosa type IV pili localizes OmcS on pili but Is deficient in Fe(III) oxide reduction and current production. Appl Environ Microbiol. 2014;80:1219–24.
Cologgi DL, Lampa-Pastirk S, Speers AM, Kelly SD, Reguera G. Extracellular reduction of uranium via Geobacter conductive pili as a protective cellular mechanism. Proc Natl Acad Sci USA. 2011;108:15248–52.
Filman DJ, Marino SF, Ward JE, Yang L, Mester Z, Bullitt E, et al. Cryo-EM reveals the structural basis of long-range electron transport in a cytochrome-based bacterial nanowire. Commun Biol. 2019;2:219.
Rotaru A-E, Shrestha PM, Liu A, Ueki T, Nevin KP, Lovley DR. Interspecies electron transfer via H2 and formate rather than direct electrical connections in co-cultures of Pelobacter carbinolicus and Geobacter sulfurreducens. Appl Environ Microbiol. 2012;78:7645–51.
Liu F, Rotaru A-E, Shrestha PM, Malvankar NS, Nevin KP, Lovley DR. Promoting direct interspecies electron transfer with activated carbon. Energy Environ Sci. 2012;5:8982–9.
Light SH, Su L, Rivera-Lugo R, Cornejo JA, Louie A, Iavarone AT, et al. A flavin-based extracellular electron transfer mechanism in diverse Gram-positive bacteria. Nature. 2018;562:140–4.
Zhang W, Zhang J, Lu Y. Stimulation of carbon nanomaterials on syntrophic oxidation of butyrate in sediment enrichments and a defined coculture. Sci Rep. 2018;8:12185.
Zhao Z, Zhang Y, Yu Q, Dang Y, Li Y, Quan X. Communities stimulated with ethanol to perform direct interspecies electron transfer for syntrophic metabolism of propionate and butyrate. Wat Res. 2016;102:475–84.
Zhao Z, Li Y, Yu Q, Zhang Y. Ferroferric oxide triggered possible direct interspecies electron transfer between Syntrophomonas and Methanosaeta to enhance waste activated sludge anaerobic digestion. Bioresour Technol. 2018;250:79–85.
Akashi H, Gojobori T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci USA. 2002;99:3695–700.
Holmes DE, Rotaru A-E, Ueki T, Shrestha PM, Ferry JG, Lovley DR. Electron and proton flux for carbon dioxide reduction in Methanosarcina barkeri during direct interspecies electron transfer. Front Microbiol. 2018;9:3109.
Coppi MV, Leang C, Sandler SJ, Lovley DR. Development of a genetic system for Geobacter sulfurreducens. Appl Environ Microbiol. 2001;67:3180–7.
Nevin KP, Kim B-C, Glaven RH, Johnson JP, Woodard TL, Methé BA, et al. Anode biofilm transcriptomics reveals outer surface components essential for high current power production in Geobacter sulfurreducens fuel cells. PLoS One. 2009;4:e5628.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Walker, D.J.F., Nevin, K.P., Holmes, D.E. et al. Syntrophus conductive pili demonstrate that common hydrogen-donating syntrophs can have a direct electron transfer option. ISME J 14, 837–846 (2020). https://doi.org/10.1038/s41396-019-0575-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-019-0575-9
This article is cited by
-
Engineering nanowires in bacteria to elucidate electron transport structural–functional relationships
Scientific Reports (2023)
-
Performance of anaerobic digestion of phenol using exogenous hydrogen and granular activated carbon and analysis of microbial community
Environmental Science and Pollution Research (2023)
-
Electromicrobiology: the ecophysiology of phylogenetically diverse electroactive microorganisms
Nature Reviews Microbiology (2022)
-
Light-driven carbon dioxide reduction to methane by Methanosarcina barkeri in an electric syntrophic coculture
The ISME Journal (2022)
-
Improving methane production from hydrogenogenic effluent with magnetic leaf biochar
Biomass Conversion and Biorefinery (2022)