Abstract
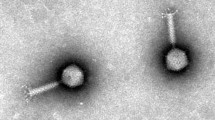
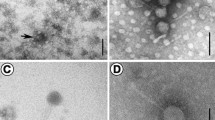
A novel jumbo bacteriophage (myovirus) is described. The lytic phage of Tenacibaculum maritimum, which is the etiological agent of tenacibaculosis in a variety of farmed marine fish worldwide, was plaque-isolated from seawater around a fish aquaculture field in Japan. The phage had an isometric head 110–120 nm in diameter, from which several 50- to 100-nm-long flexible fiber-like appendages emanate, and a 150-nm-long rigid contractile tail. The full genomes of the two representative phages (PTm1 and PTm5) were 224,680 and 226,876 bp long, respectively, both with 29.7% GC content, and the number of predicted open reading frames (ORFs) was 308 and 306, respectively. The average nucleotide sequence identity between PTm1 and PTm5 was 99.95%, indicating they are quite similar to each other. A genetic relationship was found in 15.0–16.6% of the predicted ORFs among the T. maritimum phages PTm1 and PTm5, the Tenacibaculum spp. phage pT24, and the Sphingomonas paucimobilis phage PAU. Phylogenetic analysis based on the terminase large subunit genes revealed that these four phages (PTm1, PTm5, pT24 and PAU) are more closely related than the other 10 jumbo myoviruses that have similar genome sizes. Transmission electron microscopy observations suggest that the head fibers of the T. maritimum phage function as tentacles to search and recognize the host cell surface to facilitate infection.




Similar content being viewed by others
References
Bergh Ø, Børsheim KY, Bratbak G, Heldal M (1989) High abundance of viruses found in aquatic environments. Nature 340:467–468. https://doi.org/10.1038/340467a0
Proctor LM, Fuhrman JA (1990) Viral mortality of marine bacteria and cyanobacteria. Nature 343:60–62. https://doi.org/10.1038/343060a0
Fuhrman JA, Noble RT (1995) Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol Oceanogr 40:1236–1242. https://doi.org/10.4319/lo.1995.40.7.1236
Ackermann HW, Prangishvili D (2012) Prokaryote viruses studied by electron microscopy. Arch Virol 157:1843–1849. https://doi.org/10.1007/s00705-012-1383-y
Lavigne R, Molineux IJ, Kropinski AM (2012) Caudovirales. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus Taxonomy: ninth report of the international committee on taxonomy of viruses. Elsevier Academic Press, London, pp 39–45
Guttman B, Raya R, Kutter E (2005) Basic phage biology. In: Kutter E, Sulakvelidze A (eds) Bacteriophages biology and applications. CRC Press, Boca Raton
Anderson DL, Hickman DD, Reilly BE (1966) Structure of Bacillus subtilis bacteriophage phi29 and length of phi29 deoxyribonucleic acid. J Bacteriol 91:2081–2089
Tao YZ, Olson NH, Xu W, Anderson DL, Rossmann MG, Baker TS (1998) Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell 95:431–437. https://doi.org/10.1016/S0092-8674(00)81773-0
Leonard KR, Kleinschmidt AK, Agabian-Keshishian N, Shapiro L, Maizel JV Jr (1972) Structural studies on the capsid of Caulobacter crescentus bacteriophage phiCbK. J Mol Biol 71:201–216. https://doi.org/10.1016/0022-2836(72)90346-4
Guerrero-Ferreira RC, Viollier PH, Ely B, Poindexter JS, Georgieva M, Jensen GJ, Wright ER (2011) Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc Natl Acad Sci USA 108:9963–9968. https://doi.org/10.1073/pnas.1012388108
Kalatzis PG, Bastias R, Kokkari C, Katharios P (2016) Isolation and characterization of two lytic bacteriophages, phi St2 and phi Grn1; phage therapy application for biological control of Vibrio alginolyticus in aquaculture live feeds. PLoS One. https://doi.org/10.1371/journal.pone.0151101
Frank H, Moebus K (1987) An electron microscopic study of bacteriophages from marine waters. Helgoländer Meeresunters 41:385–414. https://doi.org/10.1007/Bf02365400
Attai H, Boon M, Phillips K, Noben JP, Lavigne R, Brown PJB (2018) Larger than life: Isolation and genomic characterization of a jumbo phage that infects the bacterial plant pathogen, Agrobacterium tumefaciens. Front Microbiol 9:1861. https://doi.org/10.3389/fmicb.2018.01861
Wakabayashi H, Hikida M, Masumura K (1986) Flexibacter maritimus sp. nov., a pathogen of marine fishes. Int J Syst Bacteriol 36:396–398. https://doi.org/10.1099/00207713-36-3-396
Suzuki M, Nakagawa Y, Harayama S, Yamamoto S (2001) Phylogenetic analysis and taxonomic study of marine Cytophaga-like bacteria: proposal for Tenacibaculum gen. nov. with Tenacibaculum maritimum comb. nov. and Tenacibaculum ovolyticum comb. nov., and description of Tenacibaculum mesophilum sp. nov. and Tenacibaculum amylolyticum sp. nov. Int J Syst Evol Microbiol 51:1639–1652. https://doi.org/10.1099/00207713-51-5-1639
Bernardet JF, Nakagawa Y, Holmes B (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070. https://doi.org/10.1099/00207713-52-3-1049
Avendaño-Herrera R, Toranzo AE, Magarinos B (2006) Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: a review. Dis Aquat Org 71:255–266. https://doi.org/10.3354/dao071255
Rahman T, Suga K, Kanai K, Sugihara Y (2015) Infection kinetics of Tenacibaculum maritimum on the abraded skin of Japanese flounder Paralichthys olivaceus. Fish Pathol 50:44–52. https://doi.org/10.3147/jsfp.50.44
Pérez-Pascual D, Lunazzi A, Magdelenat G, Rouy Z, Roulet A, Lopez-Roques C, Larocque R, Barbeyron T, Gobet A, Michel G, Bernardet JF, Duchaud E (2017) The complete genome sequence of the fish pathogen Tenacibaculum maritimum provides insights into virulence mechanisms. Front Microbiol 8:1542. https://doi.org/10.3389/fmicb.2017.01542
Khoa HV, Midorikawa Y, Uchino T, Nakai T, Kato G, Kondo H, Hirono I, Labaiden M, Direkbusarakom S, Sano M (2017) Complete genome sequence of the lytic giant bacteriophage pT24 infecting Tenacibaculum spp., isolated from a shrimp culture pond. Genome Announc 5:e00081-17. https://doi.org/10.1128/genomea.00081-17
Carlson K (2005) Working with bacteriophages: common techniques and methodological approaches. In: Kutter E, Sulakvelidze A (eds) Bacteriophages biology and applications. CRC Press, Boca Raton, pp 437–494
Kawato Y, Yasuike M, Nakamura Y, Shigenobu Y, Fujiwara A, Sano M, Nakai T (2015) Complete genome sequence analysis of two Pseudomonas plecoglossicida phages, potential therapeutic agents. Appl Environ Microbiol 81:874–881. https://doi.org/10.1128/AEM.03038-14
Nakamura Y, Yasuike M, Nishiki I, Iwasaki Y, Fujiwara A, Kawato Y, Nakai T, Nagai S, Kobayashi T, Gojobori T, Ototake M (2016) V-GAP: viral genome assembly pipeline. Gene 576:676–680. https://doi.org/10.1016/j.gene.2015.10.029
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST server: Rapid annotations using subsystems technology. BMC Genom 9:75. https://doi.org/10.1186/1471-2164-9-75
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Ackermann HW, Auclair P, Basavarajappa S, Konjin HP, Savanurmath C (1994) Bacteriophages from Bombyx mori. Arch Virol 137:185–190. https://doi.org/10.1007/BF01311186
Yuan Y, Gao M (2017) Jumbo bacteriophages: an overview. Front Microbiol 8:403. https://doi.org/10.3389/fmicb.2017.00403
Crummett LT, Puxty RJ, Weihe C, Marston MF, Martiny JBH (2016) The genomic content and context of auxiliary metabolic genes in marine cyanomyoviruses. Virology 499:219–229. https://doi.org/10.1016/j.virol.2016.09.016
Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW (2005) Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol 3:e144. https://doi.org/10.1371/journal.pbio.0030144
Shen CJ, Liu YJ, Lu CP (2012) Complete genome sequence of Aeromonas hydrophila phage CC2. J Virol 86:10900. https://doi.org/10.1128/JVI.01882-12
Vincent AT, Paquet VE, Bernatchez A, Tremblay DM, Moineau S, Charette SJ (2017) Characterization and diversity of phages infecting Aeromonas salmonicida subsp. salmonicida. Sci Rep 7:7054. https://doi.org/10.1038/s41598-017-07401-7
Petrov VM, Ratnayaka S, Nolan JM, Miller ES, Karam JD (2010) Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol J 7:292. https://doi.org/10.1186/1743-422X-7-292
Kim JH, Son JS, Choi YJ, Choresca CH Jr, Shin SP, Han JE, Jun JW, Park SC (2012) Complete genome sequence and characterization of a broad-host range T4-like bacteriophage phiAS5 infecting Aeromonas salmonicida subsp salmonicida. Vet Microbiol 157:164–171. https://doi.org/10.1016/j.vetmic.2011.12.016
Lal TM, Sano M, Hatai K, Ransangan J (2016) Complete genome sequence of a giant Vibrio phage ValKK3 infecting Vibrio alginolyticus. Genom Data 8:37–38. https://doi.org/10.1016/j.gdata.2016.03.002
Luo ZH, Yu YP, Jost G, Xu W, Huang XL (2015) Complete genome sequence of a giant Vibrio bacteriophage VH7D. Mar Genom 24:293–295. https://doi.org/10.1016/j.margen.2015.10.005
Miller ES, Heidelberg JF, Eisen JA, Nelson WC, Durkin AS, Ciecko A, Feldblyum TV, White O, Paulsen IT, Nierman WC, Lee J, Szczypinski B, Fraser CM (2003) Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J Bacteriol 185:5220–5233. https://doi.org/10.1128/jb.185.17.5220-5233.2003
Matsuzaki S, Tanaka S, Koga T, Kawata T (1992) A broad-host-range vibriophage, KVP40, isolated from sea water. Microbiol Immunol 36:93–97. https://doi.org/10.1111/j.1348-0421.1992.tb01645.x
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
White RA, Suttle CA (2013) The draft genome sequence of Sphingomonas paucimobilis strain HER1398 (Proteobacteria), host to the giant PAU phage, indicates that it is a member of the genus Sphingobacterium (Bacteroidetes). Genome Announc 1:e00598-13. https://doi.org/10.1128/genomea.00598-13
Ageno M, Donelli G, Guglielmi F (1973) Structure and physico-chemical properties of bacteriophage G. II, The shape and symmetry of the capsid. Micron 4:376–403. https://doi.org/10.1016/0047-7206(73)90002-2
Suttle CA (2005) Viruses in the sea. Nature 437:356–361. https://doi.org/10.1038/nature04160
Yamada T, Satoh S, Ishikawa H, Fujiwara A, Kawasaki T, Fujie M, Ogata H (2010) A jumbo phage infecting the phytopathogen Ralstonia solanacearum defines a new lineage of the Myoviridae family. Virology 398:135–147. https://doi.org/10.1016/j.virol.2009.11.043
Labrie SJ, Samson JE, Moineau S (2010) Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. https://doi.org/10.1038/nrmicro2315
Kuznetsov YG, Chang S-C, Credaroli A, McPherson A (2013) Unique tail appendages of marine bacteriophages. Adv Microbiol 3:55–59. https://doi.org/10.4236/aim.2013.36A007
Merril CR, Scholl D, Adhya SL (2003) The prospect for bacteriophage therapy in Western medicine. Nat Rev Drug Discov 2:489–497. https://doi.org/10.1038/nrd1111
Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM (2011) Phage treatment of human infections. Bacteriophage 1:66–85. https://doi.org/10.4161/bact.1.2.15845
Balogh B, Jones JB, Iriarte FB, Momol MT (2010) Phage therapy for plant disease control. Curr Pharm Biotechnol 11:48–57. https://doi.org/10.2174/138920110790725302
Nakai T (2010) Application of bacteriophages for control of infectious diseases in aquaculture. In: Sabour PM, Griffiths MW (eds) Bacteriophages in the control of food- and waterborne pathogens. ASM Press, Washington, DC, pp 257–272
Oliveira J, Castilho F, Cunha A, Pereira MJ (2012) Bacteriophage therapy as a bacterial control strategy in aquaculture. Aquacult Int 20:879–910. https://doi.org/10.1007/s10499-012-9515-7
Acknowledgments
This study was supported by a grant-in-aid (Marine Metagenomics for Monitoring the Coastal Microbiota) from the ministry of Agriculture, Forestry and Fisheries of Japan. We thank Kanae Koike for the technical assistance with electron microscopy at Hiroshima University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Chan-Shing Lin.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kawato, Y., Istiqomah, I., Gaafar, A.Y. et al. A novel jumbo Tenacibaculum maritimum lytic phage with head-fiber-like appendages. Arch Virol 165, 303–311 (2020). https://doi.org/10.1007/s00705-019-04485-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04485-6