Abstract
Main conclusion
Certain apple cultivars accumulate to high levels in their nectar and stigma exudate an acidic chitinase III protein that can protect against pathogens including fire blight disease causing Erwinia amylovora.
Abstract
To prevent microbial infections, flower nectars and stigma exudates contain various antimicrobial compounds. Erwinia amylovora, the causing bacterium of the devastating fire blight apple disease, is the model pathogen that multiplies in flower secretions and infects through the nectaries. Although Erwinia-resistant apples are not available, certain cultivars are tolerant. It was reported that in flower infection assay, the ‘Freedom’ cultivar was Erwinia tolerant, while the ‘Jonagold’ cultivar was susceptible. We hypothesized that differences in the nectar protein compositions lead to different susceptibility. Indeed, we found that an acidic chitinase III protein (Machi3-1) selectively accumulates to very high levels in the nectar and the stigma exudate of the ‘Freedom’ cultivar. We show that three different Machi3-1 alleles exist in apple cultivars and that only the 5B-Machi3-1 allele expresses the Machi3-1 protein in the nectar and the stigma exudate. We demonstrate that the 5B-Machi3-1 allele was introgressed from the Malus floribunda 821 clone into different apple cultivars including the ‘Freedom’. Our data suggest that MYB-binding site containing repeats of the 5B-Machi3-1 promoter is responsible for the strong nectar- and stigma exudate-specific expression. As we found that in vitro, the Machi3-1 protein impairs growth and biofilm formation of Erwinia at physiological concentration, we propose that the Machi3-1 protein could partially protect 5B-Machi3-1 allele containing cultivars against Erwinia by inhibiting the multiplication and biofilm formation of the pathogen in the stigma exudate and in the nectar.
Similar content being viewed by others
Introduction
Plants secrete rewarding, sugar-rich fluids such as stigma exudates and nectar to attract pollinators (Tanveer et al. 2014). Based on their stigma, Angiosperms can be divided into two groups, plants with dry (e.g.: Arabidopsis thaliana) and wet stigma (e.g.: Malus domestica, apple and Nicotiana tabacum, tobacco). While the surface of the dry stigma is covered with a proteinaceous extracuticular layer (pellicle), the wet stigma is covered with stigma exudates (Edlund et al. 2004). This fluid secretion plays a critical role in pollen capture, hydration, growth of pollen tube and serves as a reward for the pollinators. The exudate can also be found at the intercellular spaces of the stigmatic zone and transmitting tissue in mature pistils (Rejón et al. 2014). Floral nectars are secreted by specific glands called nectaries (Heil 2011; Roy et al. 2017). Nectaries have evolved independently multiple times, and they can differ in their position, morphology, and secretion mechanism (De la Barrera and Nobel 2004). Despite these differences, nectary development is conserved in most angiosperms (Min et al. 2019). The C-lineage genes regulate the expression of the Crab Claws transcription factor, which is essential for nectary development (Bowman and Smyth 1999; Lee et al. 2005; Morel et al. 2018). Jasmonic acid (JA) is required for nectar secretion, while auxin mainly controls the volume of the nectar (Radhika et al. 2010; Bender et al. 2013; Roy et al. 2017). JA induces nectary-specific gene expression by stimulating the degradation of JAZ proteins (Kelley and Estelle 2012), thereby releasing the JAZ repressed transcriptional factor MYB305 or the homologs of MYB305 (Liu et al. 2009; Liu and Thornburg 2012; Stitz et al. 2014; Schmitt et al. 2018a). MYB305 directly and indirectly promotes the transcription of nectary-specific genes, including nectarins, genes whose proteins products are secreted into the nectar (Liu and Thornburg 2012).
The chemical composition of the stigma exudate and the nectar is relatively different, but both secretions are highly nutritious containing high levels of sugars. The stigma exudate is rich in complex sugars and proteins and contains at lower concentration free sugars, amino acids and lipids (Pusey et al. 2008). In contrast, the nectar is rich in sucrose and hexoses and free amino acids. It also contains additional components such as phenolics, secondary metabolites and nectarins (Heil 2011). The proteome of the floral nectars is relatively simple and frequently contains only a few (sometimes only one or two) dominant proteins (Roy et al. 2017). The protein profile of the stigma exudates is more complex suggesting that it is a physiologically more active extracellular fluid (Rejón et al. 2013). Both secretions are excellent medium for microbes. Microbial infection of the flower secretions is harmful as microbes can alter the chemical composition of the fluids, and mainly because plant pathogens can infect efficiently by multiplying in these nutritious fluids and then by entering into the plant through the stomata of the nectaries. Thus, it is not surprising that plants accumulate antimicrobial components including antimicrobial proteins (such as chitinases and glucanases) in these secretions (González-Teuber et al. 2009, 2010). Indeed, in the proteome of stigma exudates, the defense and stress-response proteins are the dominant GO categories (Sang et al. 2012). Flower nectars frequently contain antimicrobial proteins in very high concentrations (Zha et al. 2016; Ma et al. 2017; Nogueira et al. 2018; Schmitt et al. 2018b) or accumulate nectarins that generate antimicrobial hydrogen peroxide in the nectar (Carter et al. 2007). It was shown that nectar of wild squash is antibiotic and efficiently reduces the symptoms of the bacterial wilt (Sasu et al. 2010).
The Gram-negative bacterium Erwinia amylovora, that is one of the most devastating bacterial pathogens of apple, is the classical example of pathogens that multiply in flower secretions and infects through the nectaries (Farkas et al. 2012; Malnoy et al. 2012). Erwinia first colonizes the stigma and multiplies in the stigma exudates, then the pathogen is washed down by rain or dew into the nectar. The pathogen further multiplies in the nectar and finally enters into the plant through the stomata of the nectaries (Bubán et al. 2003). E. amylovora produces exopolysaccharides (EPS) that are involved in biofilm formation (Koczan et al. 2009). Pear fruit and apple shoot inoculation assays show that mature biofilm formation is needed for full virulence of Erwinia (Koczan et al. 2011; Piqué et al. 2015). Although Erwinia-resistant apple cultivars are not available, certain cultivars are tolerant (Gusberti et al. 2015). These cultivars are infected less frequently and develop reduced symptoms. For instance, after inoculation of the stigmas of the tolerant ‘Freedom’ and the susceptible ‘Jonagold’ cultivars, ‘Freedom’ was less infected, much less bacteria were detected on the surface of the nectaries and tissue coloring symptoms were much weaker on the ‘Freedom’ (Mihalik et al. 2004). It was assumed that the chemical composition of the ‘Freedom’ and ‘Jonagold’ nectars was identical, and proposed that the rough surface of the ‘Jonagold’ nectary was responsible for the more efficient colonization and the stronger symptoms (Mihalik et al. 2004). However, accumulating data indicate that nectarins can play important antimicrobial role (Heil 2011; Roy et al. 2017). Therefore, we wanted to test an alternative (but not mutually exclusive) hypothesis that the nectar protein profiles of the tolerant and susceptible cultivars are different. Indeed, we found that an acidic chitinase III protein (Machi3-1) accumulates to high level in the nectar and the stigma of the tolerant ‘Freedom’ cultivar, but not in the susceptible cultivars.
We show that different Machi3-1 alleles are present in ‘Freedom’ and ‘Jonagold’ cultivars and that the presence of five direct repeats in the promoter of ‘Freedom’ Machi3-1 allele is responsible for the strong nectar- and stigma-specific expression. We demonstrate that the strongly expressing Machi3-1 allele was introgressed from Malus floribunda 821 into different cultivars including ‘Freedom’. Relevantly, we found that Machi3-1 protein can inhibit the growth and biofilm formation of E. amylovora in vitro at physiological concentration. How the stigma- and nectar-specific expression of Machi3-1 could contribute to the Erwinia tolerance and in general to plant defense will be discussed.
Materials and methods
Bacterial strain and plant materials
The bacterial strain Erwinia amylovora ref T was grown in TSB (Tryptic Soy Broth) medium at 28 °C overnight. The plant materials were collected from the cultivar collection of the Research Institute for Fruitgrowing and Ornamentals (Újfehértó, Hungary) from 2005 to 2015. We used various scab-resistant and susceptible cultivars. Malus domestica Borkh. cultivars ‘Jonagold’, ‘Sampion’, ‘Golden Delicious’, ‘Gala’ ‘Idared’, ‘Redwinter’, ‘Red Rome’ and the Hungarian landrace ‘Simonffy’ are scab susceptible, while ‘Releika’, ‘Resi’, ‘Remo’, ‘Rewena’ (Germany) ‘Rajka’, ‘Selena’, ‘Topaz’, ‘Rubin’, ‘Rubinola’ (Czech Republic), ‘Hesztia’ (Hungary) are scab resistant cultivars. F1 hybrids of ‘Freedom’ × ‘Redwinter’ and ‘Freedom’ × ‘Red Rome’ derived from earlier crossbreeding program.
Collection of nectar and stigma exudate
Apple nectars were collected from field grown plants. Nectars from transgenic tobaccos, which were grown in the greenhouse, were collected in the morning (9–10 am). Nectars from the same cultivar were usually pooled and stored at − 70 °C. Stigma samples were pooled from five stigmas of the same plant. Apple stigma exudates were collected in the morning, for each sample, 30 flowers were pooled. Apple exudates were prepared as described (Pusey et al. 2008). Briefly, stigmas were submerged in water, vortexed for 2 min and then centrifuged at 13,000×g for 5 min. The supernatant was filtrated through membrane (Millex-GS; Sigma, SLGS033). Stigma exudates from tobaccos were prepared as described with some modification (Verhoeven et al. 2005). Five mature stigmas from one plant were soaked for 30 min in 200 μL of 50 mm NaAc, pH 4.5, centrifuged for 20 min, and then the supernatant was collected. Stigma exudates were frozen and kept in − 70 °C.
Nucleic acid techniques and protein extraction
Genomic DNA was purified with Quick-DNA Plant/Seed Miniprep kit (Zymo Research D6020). RNA extraction was carried out as described (Szittya et al. 2002). To purify plant protein extract, 100 mg plant tissue was homogenized with 400 µL extraction buffer (100 mM NaCl, 100 mM glycine, 10 mM EDTA, 2% SDS), incubate at 95 °C for 5 min and centrifuged. Protein concentration was measured at 280 nm.
Stain-free protein profiles and Western-blot assays
Nectars and the protein extracts were separated by stain-free 1D SDS-PAGE (Bio-Rad’s Mini PROTEAN® TGX Stain-Free™ Gels). For Western-blot assays, samples were separated by SDS-PAGE, blotted onto Amersham Protran membrane (GE Healthcare, 10600008) and hybridized with rabbit polyclonal antibody serum raised against Machi3-1. ECL Anti-Rabbit IgG Horseradish Peroxidase linked (GE Healthcare, NA934-1ML) secondary antibody was used for detection. Actin antibody (Anti-Actin Plant MerckA0480) was used for control. Chemiluminescent protein detections were conducted with ECL Western Blotting Substrate (Promega, W1001), according to the manufacturer’s instructions. Western blots were scanned with ChemiDoc MP System and analyzed with ImageLab 5.0 software (Bio-Rad).
Protein sequencing
The dominant protein band of ‘Freedom’ nectar was partially sequenced (described in details in Supplementary Materials and methods S1). Briefly, the excised protein band was in-gel digested as described (Migh et al. 2018). Peptides were analyzed by data-dependent LC–MS using a Waters Q-TOF Premier mass spectrometer online coupled to a nanoAcquity uHPLC system. Raw data were converted into a peaklist using the ProteinLynx PLGS software and the data were searched using the Batchtag Web software of the Protein Prospector search engine. As automated protein identification did not yield high confidence identifications, MS/MS data were inspected manually and high-quality MS/MS spectra were evaluated manually. Protein segments were used for degenerate PCR primer designing.
PCR cloning of the Machi3-1 gene from ‘Freedom’ and ‘Jonagold’ cultivars
Degenerated oligonucleotides (aldegf and aldegr, respectively) were designed for the predicted N-proximal ADYIWNNF and the C-proximal WNRFYDN peptide segments. cDNA was prepared from total RNA isolated from the nectary rich tissues of ‘Freedom’. PCR product was amplified, subsequently cloned and sequenced. Based on this information, specific oligonucleotides were synthesized to clone the genomic region by inverse PCR (invj1 for, invj2 for, invba1 rev, invba2 rev). The ‘Jonagold’ Machi3-1 gene was PCR amplified from the genomic DNA with the SaFreeFor and Machi3-1-stopRev primers. 5B-Machi3-1 and 5B-Machi3-1 GenBank accession numbers are BankIt2254060 5B-Machi3-1 MN496127 and BankIt2265752 2B-Machi3-1 MN496128, respectively.
RT-PCR assays
For quantitative RT-PCR, total RNAs were treated with DNase I (Thermo Fisher Scientific, EN0525), and cDNAs were transcribed using a RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, K1621). qRT-PCR was carried out with Fast Start Essential DNA Green Master Mix (Roche, 06402712001) in a Light Cycler 96 Real-Time PCR instrument (Roche). For semi-quantitative RT-PCR assays, the same cDNAs were used in conventional PCR reactions using DreamTaq Green PCR Mastermix (Thermo Fisher Scientific, K1081) in a T-Personal thermal cycler (Biometra).
Machi3-1 genotyping
The promoter regions of Machi3-1 alleles were amplified with the SaFreeFor and SaFreeRev primers and separated on 1.5% agarose gel.
Cloning of the transgenic constructs
To generate 5B-Machi3-1 and 2B-Machi3-1 transgenic constructs, the promoter and coding region of 5B-Machi3-1 and 2B-Machi3-1 genes were amplified with the 5B2BproFor and Machi3-1-stopRev primers, and then the PCR products were cloned into HindIII and BamHI cleaved Bin61S vector (Silhavy et al. 2002). To create promoter deletion constructs, promoter segments were amplified using one of the 1.2kbFor, 1kbFor, 0.9kbFor, 0.6kbFor, 0.4kbFor, 0.2kbFor forward primers and the Machi3-1stopRev reverse primer. The fragments were cloned into HindIII and HpaI cleaved 5B-Machi3-1 transformation vector to replace the original promoter regions. The constructs were sequenced. The list of primers is shown as Supplementary Table S1.
Plant transformation
Leaf disc transformation was carried out to generate transgenic N. tabacum plants (Bevan et al. 1985). Transgenic tobaccos were selected on kanamycin containing media and then the regenerants (T0 plants) were grown in the greenhouse.
Expression of recombinant proteins in Pichia pastoris, analysis of protein expression
Machi3-1 protein was expressed with Pichia Expression Kit (Invitrogen K1710-01) using pPICz vector. The protein was expressed according to the manufacturer’s protocol. The signal peptide of Machi3-1 transports the protein to the extracellular space, allowing the purification of the protein from the supernatant. The secreted protein was purified from the supernatant by precipitating with ammonium-sulfate (60% saturation). The precipitate was pelleted (25,000×g, 10 min, 4 °C), resuspended in 10 mM sodium-acetate (pH 5.0) and concentrated with Amicon Ultra-4 Centrifugal Filter Units (Merck Millipore, 10 K) ~ tenfold, reaching the final protein concentration ~ 3 mg/mL Empty pPICz vector transformant P. pastoris was grown and induced as the test strain, and then, its supernatant was similarly treated (ammonium-sulfate precipitated, centrifuged, resuspended in 10 mM Sodium-acetate and concentrated to ~ 3 mg/mL). The purified supernatant of the empty vector transformant P. pastoris was used as negative control in activity assays. SDS-PAGE assay was used to test that the background of the negative control and the purified Machi3-1 was similar.
Chitinase and lysozyme activity assay
Chitinase activity was measured by Schales’ reagent method (Ferrari et al. 2014). Colloid chitin was prepared according to Shen et al. (2010) with minor modifications. 6 g chitin was suspended in 200 mL 37% HCl and agitated overnight at 4 °C. One L of distilled water was added followed by centrifugation at 8000×g for 20 min. The pellet was washed with water till the pH reached 5.0.
Colloid chitin (at 3 mg/ml final concentration) was incubated in 200 µL of 50 mM KPO4 (pH 6.0.) with increasing amounts (50–400 ng) of Machi3-1 protein. The reactions were rotated at 30 °C for 1 h. Samples were briefly centrifuged (10 s) and 100 µL supernatant was transferred to a new tube. 100 µL Schales’ reagent (0.5 M sodium carbonate and 0.5 g/L potassium ferricyanide in water) was added and then boiled at 97 °C for 15 min. After cooling down to RT, absorbance was measured at 420 nm. Chitinase from Streptomyces griseus (Sigma, 9001-06-3) was used for positive control. Purified supernatant of the empty vector transformant P. pastoris was used as a negative control.
Lysozyme activity was measured by agar diffusion plate method. Micrococcus lysodeikticus (Merck, 4698) was used as the substrate (0.05 mg/mL). 1% agarose gel containing 1 mg M. lysodeikticus in 10 mM sodium-acetate buffer (pH 5.0.) was made (20 mL per plate). After solidification, lysozyme (Merck L6876) or Machi3-1 were loaded into the wells (4 mm diameter). Plates were incubated at 30 °C for 24 h.
In vitro Erwinia growth inhibition assay
Bacterial in vitro growth inhibition assay was carried out with minor modifications as described (Nash et al. 2006). Approximately, 102E. amylovora cells were suspended in 200 µL of 10 mM sodium-acetate buffer (pH 5.0.). The suspension was incubated without shaking for 24 h at 28 °C with different amount of purified Machi3-1 protein, or with purified supernatant of empty vector transformant P. pastoris as a negative control. Viable cells were counted by plating.
Enzymatic detachment of Erwinia biofilm
In vitro biofilm detachment assay was monitored with crystal violet staining (Koczan et al. 2009; O’Toole 2011). E. amylovora overnight culture was diluted in LB to 1:100 and 130 µL of the diluted culture was incubated at 30 °C in a 96-well TC-treated Tissue culture polystyrene plate (1 × 106 cells per well) to allow biofilm formation. After 24 h the suspension was removed, and then 130 µL of purified Machi3-1 diluted in 50 mM KPO4 buffer (pH 6.0) was added to the biofilm-covered plates. The reactions were kept at 28 °C for 3 h. Wells were washed three times with dH2O, then 150 µL 0.1% crystal violet (CV) was added. After 15 min CV was removed, then the plate was washed three times with dH2O, and dried for overnight. For quantification, 30% acetic acid was added to each well, incubated for 15 min and the OD was measured at 550 nm.
Statistics
Bacterial growth inhibition assays were repeated four times in independent experiments. Biofilm detachment experiments were performed in octuplicate and repeated three times. Comparisons between groups were done by ANOVA and Tukey test to determine P values. Spearman correlation coefficient with a P value was calculated in R Statistical Environment. Statistical significance was set at *P < 0.05, **P < 0.01 and ***P < 0.001.
Bioinformatical analysis
Sequence analysis was made by BLASTN and BLASTP softwares. Transcription factor-binding sites were predicted using PlantTFDB (Plant Transcription Factor Database) (Jin et al. 2017). Protein and DNA sequences were aligned by ClustalW method using the MegAlign program. Structural alignment and homology modeling of Hevamine as the template and the Machi3-1 protein was carried out by the SPDBV (Swiss-PdbViewer) program. Phylogeny tests were made using Bootstrap method (No. of Bootstrap Replications = 1000) and analyzed by UPGMA statistical method using the MEGA6 software.
Results
A class III chitinase-like protein accumulates in the nectar of the Erwinia-tolerant ‘Freedom’ apple cultivar
To test our hypothesis that the nectar composition of the fire blight tolerant and susceptible apple cultivars is different, nectar protein profiles of the tolerant ‘Freedom’, the susceptible ‘Jonagold’ and ‘Sampion’ cultivars were studied by 1D SDS-PAGE. None of the nectar proteins accumulated to high levels in the susceptible cultivars, while the nectar of ‘Freedom’ contained a 29 kDa-dominant protein (Fig. 1a). Although this protein was present at very high concentration (~ 50–80 ng/µL) in the ‘Freedom’ nectar, it was not detectable in the nectars of the susceptible cultivars (Fig. 1a). The analysis was repeated in 4 consecutive years with the same results, therefore, the presence of this dominant protein in the ‘Freedom’ nectar was not due to any environmental condition. The 29 kDa protein was isolated, partially sequenced, then primers were designed and inverse PCRs were conducted to clone the genomic copy of the gene from the ‘Freedom’ cultivar. The amplified region contained an 894 nucleotide (nt) long intronless coding sequence, a long (1417 nt) upstream and a short (77 nt) downstream regions. Sequence analysis revealed that the predicted ‘Freedom’ nectar protein is a class III chitinase (will be referred to as Machi3-1 for MaluschitinaseIII-1) (Fig. 1b). Class III chitinases belong to the GH18 endochitinase family (Adrangi and Faramarzi 2013). Machi3-1 is an acidic class III chitinase (calculated isoelectric point is 4.4), which shows strong sequence similarity (66.4%) to the well-characterized class III chitinases as PSC (pomegranate seed chitinase) and Hevamine (64.18%) (Terwisscha Van Scheltinga et al. 1996; Lv et al. 2011; Masuda et al. 2015). The critical catalytic amino acids and the cis-peptides (involved in chitin binding) are all conserved (Fig. 1b). Moreover, homology modeling predicts that the structure of Machi3-1 protein is highly similar to the structure of Hevamine (Fig. S1). Machi3-1 contains an N-terminal signal peptide that destines proteins towards the secretory pathways (Chung and Zeng 2017). These data suggest that the Machi3-1 protein is a functional, secretable acidic chitinase.
Machi3-1 acidic chitinase III protein accumulates to high levels in the nectar of ‘Freedom’ apple cultivar. a The nectar protein profile of ‘Jonagold’ (Jon.), ‘Sampion’ (Sam.) and ‘Freedom’ (Free.) cultivars were studied by SDS-PAGE. Note that Machi3-1 protein accumulates only in the ‘Freedom’ nectar. M. shows size marker. b Multiple sequence alignment of Machi3-1 with PSC (Pomegranate seed chitinase) and Hevamine. The N-terminal signal peptide regions were omitted from the alignment. Substrate binding cleft are shown as filled circle, while the catalytic residues are marked with open circle. The three magnesium-binding sites of PSC (3–5 amino acid/binding site) are shown by empty, light green and dark gray columns. c Machi3-1 is an active chitinase. In vitro chitinase assay was conducted with purified Machi3-1 protein and with S. griseus chitinase as positive and with supernatant from empty vector transformed strain as negative control. Note that the positive control is ~ 4 times more effective than the Machi3-1
Machi3-1 is an active chitinase
Basic class III chitinases frequently have dual chitinase and lysozyme activities, while the acidic class III chitinases have strong chitinase but only weak or no lysozyme activity (Ma et al. 2017). To characterize the Machi3-1 protein, it was expressed in P. pastoris and then it was purified from the supernatant. The chitinase and lysozyme activities of the purified Machi3-1 were tested in vitro.
To measure the chitinase activity of purified Machi3-1 protein, Schales’ procedure using colloidal chitin for a substrate was carried out (Ferrari et al. 2014). S. griseus chitinase and the supernatant of empty vector transformed P. pastoris were used as positive and negative controls, respectively. Machi3-1 proved to be a relatively efficient chitinase; its activity was ~ 25% of the S. griseus chitinase (Fig. 1c). The Machi3-1 had a barely detectable lysozyme activity in Micrococcus lysis assays (Fig. S2). Thus, we concluded that Machi3-1, like most acidic chitinase III proteins, has strong chitinase and very weak lysozyme activity.
Expression of Machi3-1 gene in ‘Freedom’ and ‘Jonagold’ cultivars
To analyze the expression pattern of Machi3-1 gene, polyclonal antibody was produced and accumulation of the Machi3-1 protein was studied in different tissues of the ‘Freedom’ and ‘Jonagold’ cultivars (Figs. 2a, S3). Confirming our earlier data, the Machi3-1 protein accumulated to very high levels in the ‘Freedom’ nectar, but it was barely detectable in the ‘Jonagold’ nectar (Fig. 2a). In the ‘Freedom’ cultivar, the Machi3-1 protein accumulated to low levels in the nectary, leaf, petal, stamen and ovary samples and to moderate levels in the stigma. In the ‘Jonagold’ cultivar, the Machi3-1 protein accumulated to low levels in all samples (Fig. 2a). The findings, that Machi3-1 has a signal peptide and that, it is abundant in the nectar indicate that Machi3-1 is secreted into the nectar. We hypothesized that the protein is also secreted from the stigma into the stigma exudates. Indeed, Machi3-1 was very abundant in the stigma exudate of the ‘Freedom’ cultivar, but it accumulated to low levels in the ‘Jonagold’ exudate (Fig. 2b). Next, we studied the expression of Machi3-1 mRNAs in the nectary, stigma and leaf samples of the two cultivars (Fig. 2c). In the ‘Freedom’ cultivar, Machi3-1 transcript expressed to very high levels in the nectary and the stigma but accumulated to low levels in the leaves. In contrast, Machi3-1 mRNA expressed to low levels in all ‘Jonagold’ samples (Fig. 2c).
Expression of Machi3-1 in ‘Freedom’ and ‘Jonagold’ apple cultivars. a, b Machi3-1 protein accumulates to moderate levels in the stigma and to high levels in the nectar and the stigma exudate of ‘Freedom’ cultivar. Western-blot assay (Wes.) was conducted to monitor the expression of Machi3-1 protein a in leaf, stigma (Sti.), nectary (Nec.), ovary (Ova.), stamen (Sta.) and sepal (Sep.) and b in the stigma and stigma exudate samples in ‘Freedom’ and ‘Jonagold’ cultivars. ‘Freedom’ nectar was used as positive control (+con.) for the Machi3-1 blot. Actin probe was used as loading control. Note that actin, which lacks signal peptide, does not accumulate in neither the nectar nor the stigma exudate. **Shows a non-specific band in the stigma exudates of both cultivars. c Expression of Machi3-1 mRNA. Quantitative RT-PCR assay was conducted to study the expression of Machi3-1 mRNA in different organs of ‘Freedom’ and ‘Jonagold’ cultivars. Machi3-1 mRNA level in the ‘Jonagold’ nectary sample was taken as one and the other expressions were calculated relative to it. Significance levels: *P value < 0.05, **P value < 0.01, ***P value < 0.001. At non-significant differences, the calculated P values are shown
Taken together, these data indicate that the Machi3-1 mRNA is efficiently expressed in the nectary and stigma tissues of the ‘Freedom’ cultivar and then, its protein product is secreted into the nectar and the stigma exudate.
The promoter regions of the ‘Freedom’ and ‘Jonagold’ Machi3-1 alleles are different
We postulated that variations in the Machi3-1 promoters are responsible for the strikingly different mRNA expression between ‘Freedom’ and ‘Jonagold’ cultivars. Therefore, the coding and the promoter regions of the Machi3-1 gene were also cloned from the ‘Jonagold’ cultivar, and then the ‘Freedom’ and ‘Jonagold’ Machi3-1 genes were compared. The nucleotide sequences of the coding regions and the predicted protein sequences are almost identical (only 4/894 nt and 2/298 amino acids are different) indicating that the coding region of Machi3-1 is well conserved in different apple cultivars (Fig. S4). However, while the promoters show strong overall similarity, the ‘Freedom’ Machi3-1 promoter is longer than the ‘Jonagold’ promoter (1417 nt and 1202 nt, respectively). The main differences were found in the middle regions of the promoter (Figs. 3a, S4–7). The ‘Freedom’ contains a 38 nt long insertion and a 15 nt long deletion relative to the ‘Jonagold’ promoter (Fig. 3a). More interestingly, both promoters contain 59–64 nt long direct repeat segments (referred to as boxes). However, the ‘Jonagold’ promoter contains only two boxes, while five boxes are present in the promoter of the ‘Freedom’ Machi3-1 gene (Figs. 3a, S4). The box1 and box5 of the ‘Freedom’ promoter resemble to the box1 and box2 of the ‘Jonagold’ promoter, respectively, while the ‘Freedom’ box2, 3 and 4 are more similar to each other (Fig. S5). Moreover, in silico studies show that four potential MYB-binding sites are present in the five-box promoter region, while the two-box region contains only one predicted MYB-binding site (Fig. S6). As MYB305 regulates nectary-specific gene expression (Liu and Thornburg 2012), this finding might explain why Machi3-1 mRNA is so abundant in the nectary of the ‘Freedom’ cultivar (also see later).
The 5B-Machi3-1 allele is required for the expression of the Machi3-1 protein in the nectar. a Non-proportional schematic representation of the three Machi3-1 alleles. White box shows the coding region. Black triangles indicate the allele-specific insertions. The differently colored boxes represent the direct repeats. The arrows show the primers that were used for PCR genotyping. Note that ps-Machi3-1 is a pseudogene. b, c Nectar expression of Machi3-1 co-segregates with the 5B-Machi3-1 ‘Freedom’ allele. b F1 progenies from ‘Freedom’ × ‘Red Rome’ crossing were PCR genotyped for the Machi3-1 alleles. ‘Freedom’ and ‘Red Rome’ genotypes (Free. and R.R., respectively) are also shown. M, DNA size marker (1.5, 1.0, 0.7 and 0.5 kb bands are shown). c Accumulation of the Machi3-1 protein in the nectars of the flowering F1 plants (others did not flower). Machi3-1 expressions were studied by stain-free gel visualization (Sta.) and by Western-blot assay (Wes.). d, e The Machi3-1 protein is abundant in the nectar of 5B-Machi3-1 allele containing apple cultivars. d Apple cultivars were PCR genotyped for Machi3-1 alleles. (Fre.-‘Freedom’, Pri.-‘Prima’, Raj.-‘Rajka’, Rel.-’Releika’, Sel.-‘Selena’, Top.-‘Topaz’, Sam.-‘Sampion’, R.W.-‘Red Winter’). e Their nectar samples of the genotyped apple cultivars were stain-free visualized (Sta.) fM. floribunda 821 contains a 5B-Machi3-1 allele
Further studies revealed that Machi3-1 was present in heterozygous form in both ‘Freedom’ and ‘Jonagold’ cultivars (Figs. 3a, S7), the second allele in both cultivars was a putative pseudogene. The promoter of the pseudogene (ps promoter for pseudogene promoter) contained two boxes. The three alleles will be referred to as 5B-Machi3-1, 2B-Machi3-1 and ps-Machi3-1, respectively (Figs. 3a, S7).
5B-Machi3-1 allele is responsible for the high Machi3-1 protein level in the nectar
We cannot exclude the possibility that Machi3-1 gene is present in multiple different copies in the ‘Freedom’ and that the Machi3-1 protein that accumulates in the nectar and the stigma exudate is not produced from 5B-Machi3-1 allele. To confirm that the 5B-Machi3-1 allele is responsible for the specific expression of Machi3-1 protein, the genotype and nectar composition of F1 hybrids from ‘Freedom’ × ‘Red Rome’ and ‘Freedom’ × ‘Red Winter’ (Free. × R.R. and Free. × R.W.) crosses and of various apple cultivars were studied.
The Machi3-1 protein was not detectable in the nectars of the ‘Red Rome’ and ‘Red Winter’ cultivars and both were homozygous for the ps-Machi3-1 pseudogene alleles (Figs. 3b, S8). The ‘Freedom’ harbors one 5B-Machi3-1 and one ps-Machi3-1 allele and contains Machi3-1 protein in the nectar. The F1 progenies segregated close to the 1:1 for 5B-Machi3-1/ps-Machi3-1 heterozygous and for ps-Machi3-1/ps-Machi3-1 homozygous plants (Free. × R.R. F1 segregated for 7:7, while Free. × R.W. F1 hybrids segregated for 11:13). Only six Free. × R. R. and eight Free. × R. W. F1 plants developed flower in the year of the study. We found that Machi3-1 protein accumulated to easily detectable levels in the nectar of all F1 progenies that inherited the ‘Freedom’ 5B-Machi3-1 allele (Figs. 3c, S8), while the progenies that inherited the ‘Freedom’ ps-Machi3-1 allele did not accumulate the protein in their nectar (stigma samples were not collected). These results indicate that the Machi3-1 gene is present in a single copy in ‘Freedom’, and that the 5B-Machi3-1 allele is required and sufficient for the intense nectar-specific (and likely stigma exudate-specific) protein expression.
Several apple cultivars were analyzed to clarify how widespread is the 5B-Machi3-1 allele and to study the association between the presence of 5B-Machi3-1 allele and the accumulation of Machi3-1 protein in the nectar (Figs. 3, S9). We found that the 5B-Machi3-1 allele is present in heterozygous or homozygous form in the genome of many (but not all) Vf scab resistance gene containing apple cultivars (Gessler and Pertot 2012), while it was not found in the cultivars that did not harbor the Vf gene (Figs. 3d, S9). The Machi3-1 protein was present in the nectar of all 5B-Machi3-1 allele containing cultivars, but it was not detectable in the nectar of cultivars lacking the 5B-Machi3-1 allele (Figs. 3e, S9). These data further confirm that the 5B-Machi3-1 allele is responsible for the accumulation of Machi3-1 protein in the nectar.
The 5B-Machi3-1 might be introgressed from the Malus floribunda 821 to different cultivars
The Vf scab resistance gene was introgressed from the M. floribunda 821 clone into various apple cultivars (Gessler and Pertot 2012). As the 5B-Machi3-1 is associated with Vf resistance gene, we postulated that the 5B-Machi3-1 allele also derived from the M. floribunda 821 ancestor (Fig. S9). We found that the M. floribunda 821 contains a 5B-Machi3-1 allele in heterozygous form (Fig. 3d), the second Machi3-1 allele has a specific promoter with three boxes (3B-Machi3-1). M. floribunda 821 5B-Machi3-1 allele is almost identical to the ‘Freedom’ 5B-Machi3-1 allele, 295/298 amino acids of the predicted proteins are identical. The promoter of M. floribunda 821 5B-Machi3-1 is also highly similar to the promoter of the ‘Freedom’ 5B-Machi3-1 gene, it contains the five boxes and all four MYB-binding sites are present (Figs. S10, S11). These data indicate that the 5B-Machi3-1 allele was introgressed from the M. floribunda 821 clone into different apple cultivars. Unfortunately, flowers were not available to test the logical assumption that Machi3-1 protein is also abundant in the nectar and the stigma exudates of M. floribunda 821.
The 5B-Machi3-1 promoter can confer nectary- and stigma-specific mRNA expression in tobacco
If the trans factors that are responsible for the nectary- and stigma-specific expression of the 5B-Machi3-1 allele in apple are also present in other dicot plants, transgenic approach can be used to identify the critical promoter regions. Transgenic tobacco lines were generated with constructs containing the promoter and the coding region of the 5B-Machi3-1 or the 2B-Machi3-1 alleles (Fig. 4a). We found that the Machi3-1 protein was easily detectable in the nectar of 4 out of 16 5B-Machi3-1 transgenic tobacco lines (although the non-expressing 5B-Machi3-1 transgenics were not further studied, we assume that many of them were silenced). In contrast, the transgenic protein could not be detected in any of the 2B-Machi3-1 (0/17 plants) transgenic tobacco nectars (Fig. 4a, b). We have also studied the accumulation of the Machi3-1 protein in the stigma and the stigma extract of two 5B-Machi3-1 plants that expressed the Machi3-1 protein in their nectars and in two 2B-Machi3-1 plants. In 5B-Machi3-1 transgenic plants, the Machi3-1 protein accumulated to high levels both in the nectar and the stigma exudate (Fig. 4b) and to low levels in the stigma (Fig. S12). In contrast, in the 2B-Machi3-1 transgenic plants, the Machi3-1 protein expressed to low levels in all three samples (Figs. 4b, S12). We have also analyzed the accumulation of the transgenic mRNAs. In 5B-Machi3-1 transgenics, the Machi3-1 transcripts expressed to low levels in leaves but accumulated to moderate levels in the nectary and to very high levels in the stigma tissues. In the 2B-Machi3-1 transgenic plants, the Machi3-1 mRNA expressed weakly in the nectary, moderately in the stigma and to very low levels in leaves (Fig. 4c). These results suggest that all trans factors that (1) are required for the nectary and stigma-specific expression of 5B-Machi3-1 promoter and those factors that (2) are important for the efficient export of the Machi3-1 protein into the nectar and the stigma exudate are all present in tobacco. Thus, the 5B-Machi3-1 promoter can be used as a valuable tool for efficient nectary- and stigma-specific expression in various dicot plants. Moreover, by utilizing the signal peptide of the Machi3-1, the transgenic proteins can be effectively secreted into the nectar and stigma exudate.
5B-Machi3-1 transgenic tobacco plants express the Machi3-1 protein in the nectar and in the stigma exudate. a The five-box promoter region of the 5B-Machi3-1 allele is required for efficient expression of the protein in the nectar of the transgenic tobaccos. Non-proportional schematic representation of the 5B-Machi3-1, the 2B-Machi3-1 (upper panel) and the various 5B-Machi3-1 promoter deletion (bottom panel) constructs that were used to generate transgenic tobacco lines. The ratio of transgenic plants expressing/non-expressing Machi3-1 protein to detectable levels in the nectar is shown at the right side (nectar Machi3-1 ±). b The Machi3-1 protein is abundant in the nectar (Nec.) and the stigma exudate (S.ex.) of the 5B-Machi3-1 transgenic tobaccos. The accumulation of the Machi3-1 protein was studied by Stain-free gel visualization (Sta.) and by Western-blot assay (Wes.). *Marks a non-specific band in the stigma exudates of non-transgenic (non-trans.) negative control as well as in the 5B-Machi3-1 and the 2B-Machi3-1 transgenic plants. cMachi3-1 mRNAs accumulates to high levels in the stigma and nectary cells of 5B-Machi3-1 transgenic plants. qRT-PCR was carried out to monitor the expression of the Machi3-1 mRNAs in different tissues of the 5B-Machi3-1 and the 2B-Machi3-1 transgenic tobaccos. For both transgenic tobaccos, 3–3 plants from two independent transformants were analyzed. mRNA level of the nectary sample of 2B-Machi3-1 was taken as 1 and the other expressions were calculated relative to it. Significance levels: *P value < 0.05, **P value < 0.01, ***P value < 0.001. Non-significant P values are shown
The five-box region of the Machi3-1 promoter is required for efficient expression
Multiplication of a repeat region in a promoter can dramatically increase transcriptional activity (Espley et al. 2009). We assumed that the five-box-containing 5B-Machi3-1 promoter is responsible for the strong nectary-, and stigma-specific expression of Machi3-1 mRNA and indirectly for the nectar- and stigma exudate-specific accumulation of Machi3-1 protein. To test this assumption, plants were transformed with several deletion constructs generated from the 5B-Machi3-1 plasmid (Fig. 4a, bottom panel), and then we studied the accumulation of the Machi3-1 protein in the nectars of the transformants (stigma samples were not collected in this experiment). 15/48 plants expressed Machi3-1 protein in the nectar (Fig. 4a) when the five-box region was present in the promoter (combining the results of 1.2, 1.0 and 0.9 constructs). In contrast, 1 out of 52 plants accumulated Machi3-1 protein in the nectar at detectable levels when constructs lacking the five boxes were used (we combined the results of 0.6, 0.4 and 0.2 constructs, see Fig. 4a). These results indicate that the five-box region is essential for the efficient nectary-specific expression of 5B-Machi3-1 promoter.
The MYB305 and 5B-Machi3-1 mRNA expressions are positively correlated
We found that four potential MYB-binding sites are present in the five-box region of the 5B-Machi3-1 allele but only one site is present in the two-box promoter region and that the five-box promoter region is required for the efficient nectary- and stigma-specific expression of the 5B-Machi3-1 mRNA in both apple and tobacco (Figs. 3, 4). As the MYB305 directs the nectary-specific expression of many genes in tobacco (Liu et al. 2009), we hypothesized that in tobacco the MYB305 and in apple, the MdMYB305 (the putative apple orthologue of MYB305, gene: MDP0000344978, Fig. S13a) transcription factors stimulate the expression of 5B-Machi3-1 in the nectary cells. We hypothesized that MYB305 and MdMYB305 also promote the expression of the 5B-Machi3-1 in the stigma cells. If these assumptions are correct, MdMYB305 and MYB305 mRNAs are co-expressed with 5B-Machi3-1 (but not 2B-Machi3-1) transcripts in apple and in transgenic tobacco, respectively. To test this, qRT-PCR and semi-qRT-PCR assays were conducted to monitor the expression of MdMYB305 in ‘Freedom’ and ‘Jonagold’ cultivars, and the expression of MYB305 in transgenic tobacco lines (Fig. 5a, b). Then, the expression of MdMYB305 and MYB305 and the 5B-Machi3-1 and 2B-Machi3-1 mRNAs were compared (Figs. 2c, 4c, 5). The MdMYB305 mRNA expressed similarly in both ‘Freedom’ and ‘Jonagold’ cultivars, it was abundant in the nectary and stigma samples but was barely detectable in leaves (Fig. 5a). Thus, MdMYB305 and 5B-Machi3-1 mRNAs (but not 2B-Machi3-1 transcripts) are co-expressed, both transcripts are abundant in the nectary and the stigma cells (Figs. 4, 5). To analyze the connections between Machi3-1 and MdMYB305 mRNA levels, Spearman correlation coefficients and P values were calculated for nectary and stigma samples of ‘Freedom’ and ‘Jonagold’ cultivars (Fig. 5c). Relevantly, a strong positive correlation was observed in the stigma and nectary tissues of the ‘Freedom’ cultivar between the Machi3-1 and MdMYB305 mRNA expressions (R = 0.916 and 0.738, respectively), while correlation was not found in these tissues in the ‘Jonagold’ cultivar (Fig. 5c). To examine if 5B-Machi3-1 and MYB305 transcripts are also co-expressed in transgenic tobacco, MYB305 mRNA levels were studied in the stigma, nectary and leaf tissues of 5B-Machi3-1 and 2B-Machi3-1 transgenic tobaccos (Fig. 5b). As expected, MYB305 mRNA expressed similarly in both types of transgenic tobacco lines. In accordance with previous report (Liu et al. 2009), we found that the MYB305 mRNAs were abundant in the nectary samples (Fig. 5b). Moreover, we show that MYB305 mRNAs also expressed to high levels in stigma and to low levels in the leaf cells. Relevantly, we found that the 5B-Machi3-1 transgenic mRNA also accumulated to very high levels in the stigma and to moderate levels in the nectary and that the 2B-Machi3-1 mRNA expressed to much lower levels in these tissues (Fig. 4c). Correlation analysis suggests that MYB305 and 5B-Machi3-1 transgenic mRNA expressions are also positively correlated in the stigma and the nectary of transgenic tobaccos (Fig. 5d). The correlation was strong for the nectary (R = 0.812, P = 0.024) and less convincing for the stigma (R = 0.526, P = 0.14). The 2B-Machi3-1 mRNA expression did not correlate with either the MdMYB305 or the MYB305 transcript. These results that 5B-Machi3-1 expressions positively correlate with the expressions of MdMYB305 and MYB305 are consistent with the model that these transcription factors promote the expression of 5B-Machi3-1 in the nectary and stigma cells in apple and transgenic tobaccos (also see Discussion). Unfortunately, we failed transiently overexpress MYB305 protein in N. benthamiana, therefore, we could not directly test whether MYB305 activates 5B-Machi3-1 expression.
5B-Machi3-1 and MYB305 expressions show strong correlation. a, b qRT-PCR was carried out to monitor the expression of MdMYB305 and MYB305 transcripts. aMdMYB305 is highly expressed in the nectary and the stigma but not in the leaf cells of the ‘Freedom’ and the ‘Jonagold’ cultivars. mRNA level of the stigma sample of ‘Freedom’ was taken as 1 and the other expressions were calculated relative to it. bMYB305 mRNAs levels in different tissues of the 5B-Machi3-1 and the 2B-Machi3-1 transgenic tobaccos. mRNA of the stigma sample of 5B-Machi3-1 was taken as 1 and the other expressions were calculated relative to it. c, d To assess the connections between MYB305 and Machi3-1 mRNA expressions, Spearman correlation coefficients (R) and the corresponding P values were calculated from the qRT-PCR results shown at A and B. panels. c5B-Machi3-1 and MdMYB305 mRNA expressions positively correlate in both nectary and stigma samples of ‘Freedom’ cultivar. d5B-Machi3-1 and MYB305 mRNA expressions positively correlated in the nectary and stigma samples in the 5B-Machi3-1 transgenic tobacco
Machi3-1 inhibits growth and biofilm formation of E. amylovora in vitro
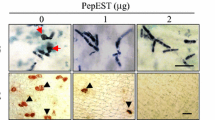
The Machi3-1 chitinase accumulated to high levels in the nectar and the stigma exudates of the ‘Freedom’ apple cultivar. As chitinases are antimicrobial proteins and because during infection, E. amylovora multiplies in the nectar and the stigma exudate, we hypothesize that Machi3-1 can partially protect ‘Freedom’ from E. amylovora. As a first step towards understanding the role of Machi3-1 in defense against E. amylovora, we tested the effect of Machi3-1 on the growth and biofilm formation of the pathogen in vitro. Erwinia cultures were incubated with Machi3-1 protein purified from the supernatant of Machi3-1 expressing P. pastoris. We found that at high-concentration (40–80 ng/µL) Machi3-1 significantly reduced the growth of E. amylovora (Fig. 6a). Relevantly, the Machi3-1 protein is present in the nectar of ‘Freedom’ cultivar in similar ~ 50–80 ng/µL concentration (Fig. S2b). As a negative control, Erwinia cultures were incubated with supernatant of empty vector transformant P. pastoris.
Machi3-1 has an antibacterial effect against E. amylovora. a Machi3-1 inhibits the growth of E. amylovora. Erwinia culture was treated with Machi3-1 protein that was purified from the supernatant of Machi3-1 expressing Pichia and with supernatant from empty vector transformed Pichia as a negative control. b Biofilm detachment assay shows that Machi3-1 impairs biofilm formation. Note that, although at 80 ng/µL concentration even the negative control has a mild activity, this effect can be separated from the effect of Machi3-1, because Machi3-1 inhibited significantly more effectively the biofilm formation than the negative control and at lower concentrations only the Machi3-1 inhibited the biofilm formation. Significance levels: *P value < 0.05, ***P value < 0.001. c Model of the regulation of 5B-Machi3-1 expression (for details, see the main text)
Biofilm formation is required for successful Erwinia infection (Koczan et al. 2011). As chitinases can impair biofilm formation (Chung et al. 2014), we wanted to study if Machi3-1 modifies biofilm formation of E. amylovora. Pre-formed E. amylovora biofilm was treated with purified Machi3-1 protein, and then biofilm detachment was quantified by crystal violet staining assay (O’Toole 2011). As Fig. 6b shows, the Machi3-1 efficiently detached the pre-formed E. amylovora biofilm at physiological concentration. Taken together, the Machi3-1 acidic chitinase III protein efficiently impairs the growth and biofilm formation of E. amylovora in vitro at physiological concentration.
Discussion
Here, we show that the 5B-Machi3-1 allele encodes an acidic chitinase III protein that accumulates to very high levels in the stigma exudate and the nectar, the primary niches of the Erwinia infection. As the Machi3-1 protein inhibits the growth and biofilm formation of Erwinia at physiological concentration in vitro, we hypothesize that the Machi3-1 protein could partially protect apple against Erwinia infection and defend nutritious flower secretions from microbial infections. We propose that the MYB305 protein homologs activate the transcription of the 5B-Machi3-1 mRNAs in the nectary and stigma cells and then the exported protein products accumulate in the nectar and stigma exudate.
Machi3-1 protein might interfere with Erwinia infection at two different steps
We found that the Machi3-1 acidic chitinase III protein is present at very high concentration in the nectar and the stigma exudate of the 5B-Machi3-1 allele containing apple cultivars (Fig. 2). As chitinases have wide spectrum antimicrobial effect (Cletus et al. 2013), the high concentration of Machi3-1 protein could effectively protect the nectar and the stigma exudate from various microbial infection. Thus, it can keep the optimal composition of these important fluid secretions, thereby enhancing the efficiency of pollination and fertilization. We propose that the high level of Machi3-1 in the stigma exudate and the nectar could partially protect 5B-Machi3-1 allele containing apple cultivars against Erwinia. During apple infection, Erwinia propagates first in the stigma exudate, then in the nectar and finally enters into the plant through the stomata of the nectary (Bubán et al. 2003; Farkas et al. 2012). As biofilm formation is critical for the fruit and shoot infection of Erwinia (Koczan et al. 2011), it is likely that biofilm formation is also required for efficient flower infection. In vitro, the Machi3-1 protein inhibits growth and biofilm formation of Erwinia at a concentration in which it is present in the nectar of 5B-Machi3-1 allele containing apple cultivars (Figs. 6, S2). These findings suggest that the Machi3-1 protein can interfere with the propagation and infection of Erwinia in the nectar of the 5B-Machi3-1 allele containing apples. Unfortunately we cannot directly measure the concentration of Machi3-1 protein in the stigma exudate. However, as Machi3-1 is one of the most abundant proteins in the ‘Freedom’ stigma exudate (Fig. S14.), it is conceivable that the Machi3-1 protein is also present in inhibitory concentration in the exudate. Thus, we propose that accumulation of Machi3-1 protein could protect apples from Erwinia infection by forming two barriers, it interferes with the replication and biofilm formation of the Erwinia in the stigma exudate as well as in the nectar. This model would explain why the 5B-Machi3-1 expressing ‘Freedom’ was more tolerant in flower inoculation assay against Erwinia than the non-expressing ‘Jonagold’ cultivar. Importantly, if 5B-Machi3-1 contributes to the Erwinia resistance, this effect is not detectable in the frequently used shoot inoculation assays.
The molecular mechanism how Machi3-1 inhibits the growth and biofilm formation of Erwinia is not known. Chitinase treatment modulated the bacterial adhesion and inhibited the biofilm formation of the Gram-negative Francisella novicida. It was hypothesized that chitinases alter the EPS composition of F. novicida thereby impairing the biofilm formation and altering adhesion (Chung et al. 2014). As amylovoran and levan EPSs play a critical role in biofilm formation and pathogenicity of Erwinia (Koczan et al. 2009), we speculate that Machi3-1 modulates the EPS of Erwinia, which results in slower growth and inefficient biofilm formation.
MYB305 could play an important role in the regulation of 5B-Machi3-1
We found that the Machi3-1 gene is present in at least three different forms in various apple cultivars, one is a pseudogene, while the two other alleles (5B-Machi3-1 and 2B-Machi3-1) encode very similar proteins (Fig. 3). The regulation of the two active alleles is markedly different. The 2B-Machi3-1 allele expresses to low levels in all tissues, while the 5B-Machi3-1 mRNAs shows intense expression in the nectary and the stigma (Fig. 2). The Machi3-1 protein contains a signal peptide that facilitates its export into the nectar and the stigma exudate explaining why the 5B-Machi3-1 mRNA is expressed in the nectary and the stigma tissues, while its protein product accumulates in the secretions.
The 5B-Machi3-1 gene is similarly expressed in apple and in transgenic tobacco suggesting that the transcription factors that are required for the specific expression of 5B-Machi3-1 are conserved in dicots (Figs. 2, 4). Our segregation, association and transgenic assays demonstrate that the five-box region of the 5B-Machi3-1 promoter is required for the nectary- and stigma-specific transcript expressions (Figs. 3, 4). We identified four potential MYB-binding sites on the five-box region of the 5B-Machi3-1 allele, but only one on the 2B promoter (Fig. S6). MYB305 transcription factor homologs play a critical role in the nectary-specific expression in tobacco, snapdragon and Arabidopsis (Roy et al. 2017). MYB305 is activated by JA, then it binds to the promoters of many nectary-specific genes and promotes their transcription. We found that the 5B-Machi3-1 and MdMYB305 mRNAs are co-expressed in apple, both transcripts accumulated in the nectary and stigma cells. The expression of these transcripts shows strong positive correlation (Fig. 5), higher MdMYB305 transcript level was associated with almost proportionally increased 5B-Machi3-1 mRNA expressions. Furthermore, the 5B-Machi3-1 and MYB305 transcript levels also show positive correlation in the nectary (and less convincing in the stigma) cells of transgenic tobacco (Figs. 5, S13). These data support the model that in both plants, the high MYB305 transcript level results in high MYB305 transcription factor concentration, which leads to the intense expression of 5B-Machi3-1 mRNAs. We hypothesize that in apple as well as in tobacco, the MYB305 level is increased (presumably due to enhanced JA concentrations) in the nectary and the stigma, thus in these tissues, MYB305 can bind to the five-box region of 5B-Machi3-1 gene and stimulate its transcription (Fig. 6c). In contrast, MYB305 fails to activate the transcription of the 2B-Machi3-1 gene. In line with this latter element of our model, we found that 2B-Machi3-1 mRNAs expressed to low levels in the 2B-Machi3-1 allele containing apple cultivars and the 2B-Machi3-1 transgenic tobaccos and its expression was independent from the expression of the MYB305 transcription factor. A recent RNA-seq. experiment also showed that in the stigma (nectary was not studied) of the 2B-Machi3-1 containing ‘Golden Delicious’ apple cultivar (SRA-NCBI: SRP125281 study), the MdMYB305 mRNA was abundant, while the 2B-Machi3-1 transcript accumulated to low levels (Fig. S13.).
Interestingly, the strong nectary and stigma expression of MYB305 is not restricted to the plants having wet stigma such as tobacco or apple. The Arabidopsis homolog MYB21 is also expressed to high levels in both nectary and in the papilla cells of the dry stigma (Osaka et al. 2013).
Author contribution statement
AK designed and performed the experiments and participated in the writing of the manuscript, TT and TL conducted apple flower studies, LD initiated the program, performed various assays and participated in the writing of the manuscript, ZD carried out protein sequencing analysis, DS designed experiments, supported the program and participated in the writing of the manuscript, ZK conducted association assays, GD designed and performed experiments.
Abbreviations
- EPS:
-
Exopolysaccharide
- JA:
-
Jasmonic acid
- Machi3-1:
-
Malus chitinaseIII-1
References
Adrangi S, Faramarzi MA (2013) From bacteria to human: a journey into the world of chitinases. Biotechnol Adv 31:1786–1795
Bender RL, Fekete ML, Klinkenberg PM et al (2013) PIN6 is required for nectary auxin response and short stamen development. Plant J 74:893–904
Bevan MW, Mason SE, Goelet P (1985) Expression of tobacco mosaic virus coat protein by a cauliflower mosaic virus promoter in plants transformed by Agrobacterium. EMBO J 4:1921–1926
Bowman JL, Smyth DR (1999) CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126:2387–2396
Bubán T, Orosz-Kovács Z, Farkas Á (2003) The nectary as the primary site of infection by Erwinia amylovora (Burr.). Plant Syst Evol 238:183–194
Carter C, Healy R, O’Tool NM, Naqvi SMS, Ren G, Park S, Beattie GA, Horner HT, Thornburg RW (2007) Tobacco nectaries express a novel NADPH oxidase implicated in the defense of floral reproductive tissues against microorganisms. Plant Physiol 143:389–399
Chung KP, Zeng Y (2017) An overview of protein secretion in plant cells. Methods Mol Biol 1662:19–32
Chung M-C, Dean S, Marakasova ES, Nwabueze AO, van Hoek ML (2014) Chitinases are negative regulators of Francisella novicida biofilms. PLoS One 9(3):e93119
Cletus J, Balasubramanian V, Vashisht D, Sakthivel N (2013) Transgenic expression of plant chitinases to enhance disease resistance. Biotechnol Lett 35:1719–1732
De la Barrera E, Nobel PS (2004) Nectar: properties, floral aspects, and speculations on origin. Trends Plant Sci 9:65–69
Edlund AF, Swanson R, Preuss D (2004) Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell 16(Suppl):S84–S97
Espley RV, Brendolise C, Chagne D et al (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21:168–183
Farkas Á, Mihalik E, Dorgai L, Bubán T (2012) Floral traits affecting fire blight infection and management. Trees 26:1–20
Ferrari AR, Gaber Y, Fraaije MW (2014) A fast, sensitive and easy colorimetric assay for chitinase and cellulase activity detection. Biotechnol Biofuels 7(1):37
Gessler C, Pertot I (2012) Vf scab resistance of Malus. Trees 26:1–14
González-Teuber M, Eilmus S, Muck A, Svatos A, Heil M (2009) Pathogenesis-related proteins protect extrafloral nectar from microbial infestation. Plant J 58:464–473
González-Teuber M, Pozo MJ, Muck A, Svatos A, Adame-Alvarez RM, Heil M (2010) Glucanases and chitinases as causal agents in the protection of Acacia extrafloral nectar from infestation by phytopathogens. Plant Physiol 152:1705–1715
Gusberti M, Klemm U, Meier MS, Maurhofer M, Hunger-Glaser I (2015) Fire blight control: the struggle goes on a comparison of different fire blight control methods in Switzerland with respect to biosafety, efficacy and durability. Int J Environ Res Public Health 12:11422–11447
Heil M (2011) Nectar: generation, regulation and ecological functions. Trends Plant Sci 16:191–200
Jin J, Tian F, Yang D-C, Meng Y-Q, Kong L, Luo J, Gao G (2017) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res 45:D1040–D1045
Kelley DR, Estelle M (2012) Ubiquitin-mediated control of plant hormone signaling. Plant Physiol 160:47–55
Koczan JM, McGrath MJ, Zhao Y, Sundin GW (2009) Contribution of Erwinia amylovora exopolysaccharides amylovoran and levan to biofilm formation: implications in pathogenicity. Phytopathology 99:1237–1244
Koczan JM, Lenneman BR, McGrath MJ, Sundin GW (2011) Cell surface attachment structures contribute to biofilm formation and xylem colonization by Erwinia amylovora. Appl Environ Microbiol 77:7031–7039
Lee J-Y, Baum SF, Oh S-H, Jiang C-Z, Chen J-C, Bowman JL (2005) Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development 132:5021–5032
Liu G, Thornburg RW (2012) Knockdown of MYB305 disrupts nectary starch metabolism and floral nectar production. Plant J 70:377–388
Liu G, Ren G, Guirgis A, Thornburg RW (2009) The MYB305 transcription factor regulates expression of Nectarin genes in the ornamental tobacco floral nectary. Plant Cell 21:2672–2687
Lv C, Masuda T, Yang H, Sun L, Zhao G (2011) High-capacity calcium-binding chitinase III from pomegranate seeds (Punica granatum Linn.) is located in amyloplasts. Plant Signal Behav 6:1963–1965
Ma XL, Milne RI, Zhou HX, Fang JY, Zha HG (2017) Floral nectar of the obligate outcrossing Canavalia gladiata (Jacq.) DC. (Fabaceae) contains only one predominant protein, a class III acidic chitinase. Plant Biol (Stuttg) 19:749–759
Malnoy M, Martens S, Norelli JL, Barny M-A, Sundin GW, Smits THM, Duffy B (2012) Fire blight: applied genomic insights of the pathogen and host. Annu Rev Phytopathol 50:475–494
Masuda T, Zhao G, Mikami B (2015) Crystal structure of class III chitinase from pomegranate provides the insight into its metal storage capacity. Biosci Biotechnol Biochem 79:45–50
Migh E, Götz T, Földi I et al (2018) Microtubule organization in presynaptic boutons relies on the formin DAAM. Development 145(6):dev158519
Mihalik E, Radvánszky A, Dorgai L, Bubán T (2004) Study of Erwinia amylovora colonization and migration on blossoms of susceptible and tolerant apple cultivars. Int J Horticult Sci 10(2):15–19
Min Y, Bunn JI, Kramer EM (2019) Homologs of the STYLISH gene family control nectary development in Aquilegia. New Phytol 221:1090–1100
Morel P, Heijmans K, Ament K, Chopy M, Trehin C, Chambrier P, Rodrigues Bento S, Bimbo A, Vandenbussche M (2018) The floral C-lineage genes trigger nectary development in Petunia and Arabidopsis. Plant Cell 30:2020–2037
Nash JA, Ballard TNS, Weaver TE, Akinbi HT (2006) The peptidoglycan-degrading property of lysozyme is not required for bactericidal activity in vivo. J Immunol 177:519–526
Nogueira FCS, Farias ARB, Teixeira FM, Domont GB, Campos FAP (2018) Common features between the proteomes of floral and extrafloral nectar from the castor plant (Ricinus communis) and the proteomes of exudates from carnivorous plants. Front Plant Sci 9:549
O’Toole GA (2011) Microtiter dish biofilm formation assay. J Vis Exp. https://doi.org/10.3791/2437
Osaka M, Matsuda T, Sakazono S et al (2013) Cell type-specific transcriptome of Brassicaceae stigmatic papilla cells from a combination of laser microdissection and RNA sequencing. Plant Cell Physiol 54:1894–1906
Piqué N, Miñana-Galbis D, Merino S, Tomás JM (2015) Virulence factors of Erwinia amylovora: a review. Int J Mol Sci 16:12836–12854
Pusey PL, Rudell DR, Curry EA, Mattheis JP (2008) Characterization of stigma exudates in aqueous extracts from apple and pear flowers. HortSci 43:1471–1478
Radhika V, Kost C, Boland W, Heil M (2010) The role of jasmonates in floral nectar secretion. PLoS One 5:e9265
Rejón JD, Delalande F, Schaeffer-Reiss C, Carapito C, Zienkiewicz K, de Dios Alché J, Rodríguez-García MI, Van Dorsselaer A, Castro AJ (2013) Proteomics profiling reveals novel proteins and functions of the plant stigma exudate. J Exp Bot 64:5695–5705
Rejón JD, Delalande F, Schaeffer-Reiss C, Carapito C, Zienkiewicz K, Alché JDD, Rodríguez-García MI, Van Dorsselaer A, Castro AJ (2014) The plant stigma exudate. A biochemically active extracellular environment for pollen germination? Plant Signal Behav 9:e28274
Roy R, Schmitt AJ, Thomas JB, Carter CJ (2017) Nectar biology: from molecules to ecosystems. Plant Sci 262:148–164
Sang YL, Xu M, Ma FF, Chen H, Xu XH, Gao X-Q, Zhang XS (2012) Comparative proteomic analysis reveals similar and distinct features of proteins in dry and wet stigmas. Proteomics 12:1983–1998
Sasu MA, Wall KL, Stephenson AG (2010) Antimicrobial nectar inhibits a florally transmitted pathogen of a wild Cucurbita pepo (Cucurbitaceae). Am J Bot 97:1025–1030
Schmitt AJ, Roy R, Klinkenberg PM, Jia M, Carter CJ (2018a) The octadecanoid pathway, but not COI1, is required for nectar secretion in Arabidopsis thaliana. Front Plant Sci 9:1060
Schmitt AJ, Sathoff AE, Holl C, Bauer B, Samac DA, Carter CJ (2018b) The major nectar protein of Brassica rapa is a non-specific lipid transfer protein, BrLTP2.1, with strong antifungal activity. J Exp Bot 69:5587–5597
Shen C-R, Chen Y-S, Yang C-J, Chen J-K, Liu C-L (2010) Colloid chitin azure is a dispersible, low-cost substrate for chitinase measurements in a sensitive, fast, reproducible assay. J Biomol Screen 15:213–217
Silhavy D, Molnár A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyán J (2002) A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J 21:3070–3080
Stitz M, Hartl M, Baldwin IT, Gaquerel E (2014) Jasmonoyl-l-isoleucine coordinates metabolic networks required for anthesis and floral attractant emission in wild tobacco (Nicotiana attenuata). Plant Cell 26:3964–3983
Szittya G, Molnár A, Silhavy D, Hornyik C, Burgyán J (2002) Short defective interfering RNAs of tombus viruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14:359–372
Tanveer T, Shaheen K, Parveen S, Kazi AG, Ahmad P (2014) Plant secretomics: identification, isolation, and biological significance under environmental stress. Plant Signal Behav 9:e29426
Terwisscha Van Scheltinga AC, Hennig M, Dijkstra BW (1996) The 1.8 Å resolution structure of hevamine, a plant chitinase/lysozyme, and analysis of the conserved sequence and structure motifs of glycosyl hydrolase family 18. J Mol Biol 262:243–257
Verhoeven T, Feron R, Wolters-Arts M, Edqvist J, Gerats T, Derksen J, Mariani C (2005) STIG1 controls exudate secretion in the pistil of petunia and tobacco. Plant Physiol 138:153–160
Zha H-GG, Milne RI, Zhou H-X, Chen X-Y, Sun H (2016) Identification and cloning of class II and III chitinases from alkaline floral nectar of Rhododendron irroratum, Ericaceae. Planta 244:805–818
Acknowledgements
Open access funding provided by MTA Biological Research Center (MTA SZBK). We are grateful to M. Peline Toth, M. Csanyi and J. Nadudvarine Novak for transformations and various technical assistances. We thank A. Kerekes (Agricultural Biotechnology Institute) for the anti-Machi3-1 antibody and A. Auber (Agricultural Biotechnology Institute) for his help with the bioinformatical studies and Z. K. Varga for his help in statistical analyses. We are especially grateful to E. Van de Weg (Wageningen, Netherlands) for the M. floribunda 821 DNA samples. We thank A. Hegedus (Szent Istvan University, Hungary) for the useful comments about the manuscript. Research was supported by grants from the OTKA (K129177, K116963). A. Kurilla is a graduate student of ELTE “Classical and Molecular Genetics” Ph.D. program.
Author information
Authors and Affiliations
Corresponding author
Additional information
To the memory of Dr. Tamás Bubán.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kurilla, A., Toth, T., Dorgai, L. et al. Nectar- and stigma exudate-specific expression of an acidic chitinase could partially protect certain apple cultivars against fire blight disease. Planta 251, 20 (2020). https://doi.org/10.1007/s00425-019-03303-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-019-03303-2