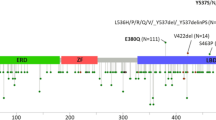
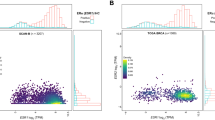
Abstract
Purpose
Triple-negative breast cancer (TNBC)/basal-like breast cancer (BLBC) is a highly aggressive form of breast cancer. We previously reported that a small molecule agonist ligand for the orphan nuclear receptor estrogen-related receptor beta (ERRβ or ESRRB) has growth inhibitory and anti-mitotic activity in TNBC cell lines. In this study, we evaluate the association of ESRRB mRNA, copy number levels, and protein expression with demographic, clinicopathological, and gene expression features in breast tumor clinical specimens.
Methods
ESRRB mRNA-level expression and clinical associations were analyzed using RNAseq data. Array-based comparative genomic hybridization determined ESRRB copy number in African-American and Caucasian women. Transcription factor activity was measured using promoter–reporter luciferase assays in TNBC cell lines. Semi-automatic quantification of immunohistochemistry measured ERRβ protein expression on a 150-patient tissue microarray series.
Results
ESRRB mRNA expression is significantly lower in TNBC/BLBC versus other breast cancer subtypes. There is no evidence of ESRRB copy number loss. ESRRB mRNA expression is correlated with the expression of genes associated with neuroactive ligand–receptor interaction, metabolic pathways, and deafness. These genes contain G/C-rich transcription factor binding motifs. The ESRRB message is alternatively spliced into three isoforms, which we show have different transcription factor activity in basal-like versus other TNBC cell lines. We further show that the ERRβ2 and ERRβsf isoforms are broadly expressed in breast tumors at the protein level.
Conclusions
Decreased ESRRB mRNA expression and distinct patterns of ERRβ isoform subcellular localization and transcription factor activity are key features in TNBC/BLBC.








Similar content being viewed by others
Abbreviations
- TNBC:
-
Triple-negative breast cancer
- BLBC:
-
Basal-like breast cancer
- AA:
-
African-American
- ESRRB:
-
Estrogen-related receptor beta
- IHC:
-
Immunohistochemistry
- ER:
-
Estrogen receptor
- PR:
-
Progesterone receptor
- HER2:
-
Human epidermal growth factor two
- CW:
-
Caucasian/White
- NR:
-
Nuclear receptor(s)
- ONR:
-
Orphan nuclear receptor
- ERR:
-
Estrogen-related receptors
- OS:
-
Overall survival
- SCAN-B:
-
Sweden Cancerome Analysis Network-Breast
- FPKM:
-
Fragments per kilobase of transcript per million mapped reads
- ESR1:
-
Estrogen receptor
- NHG:
-
Nottingham grade
- NTN:
-
Non-triple-negative breast cancer
- aCGH:
-
Array comparative genomic hybridization
- AFR:
-
African descent
- AMR:
-
Ad mixed American
- DEGs:
-
Differentially expressed genes
- BL2:
-
Basal-like 2
- LAR:
-
Luminal androgen receptor
- ML:
-
Mesenchymal-like
- ERRE:
-
Estrogen-related response element
- SP1:
-
Specificity-protein-1
- TMA:
-
Tissue microarray
References
American Cancer Society (2017) Breast cancer facts and figures 2017–2018. American Cancer Society I, Atlanta
Anders CK, Abramson V, Tan T, Dent R (2016) The evolution of triple-negative breast cancer: from biology to novel therapeutics. Am Soc Clin Oncol Educ Book 35:34–42
Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z et al (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27(8):1160–1167
Wallden B, Storhoff J, Nielsen T, Dowidar N, Schaper C, Ferree S, Liu S, Leung S, Geiss G, Snider J et al (2015) Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genomics 8:54
Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO (2007) Prognostic markers in triple-negative breast cancer. Cancer 109(1):25–32
Feng Y, Spezia M, Huang S, Yuan C, Zeng Z, Zhang L, Ji X, Liu W, Huang B, Luo W et al (2018) Breast cancer development and progression: risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis 5(2):77–106
Scott LC, Mobley LR, Kuo TM, Il’yasova D (2019) Update on triple-negative breast cancer disparities for the United States: a population-based study from the United States Cancer Statistics database, 2010 through 2014. Cancer. https://doi.org/10.1002/cncr.32207
Costa RLB, Gradishar WJ (2017) Triple-negative breast cancer: current practice and future directions. J Oncol Pract 13(5):301–303
Mullican SE, Dispirito JR, Lazar MA (2013) The orphan nuclear receptors at their 25-year reunion. J Mol Endocrinol 51(3):T115–T140
Giguère V, Yang N, Segui P, Evans RM (1988) Identification of a new class of steroid hormone receptors. Nature 331(6151):91–94
Heckler MM, Zeleke TZ, Divekar SD, Fernandez AI, Tiek DM, Woodrick J, Farzanegan A, Roy R, Üren A, Mueller SC et al (2016) Antimitotic activity of DY131 and the estrogen-related receptor beta 2 (ERRβ2) splice variant in breast cancer. Oncotarget 7(30):47201–47220
Long MD, Campbell MJ (2015) Pan-cancer analyses of the nuclear receptor superfamily. Nucl Recept Res. https://doi.org/10.11131/2015/101182
Garattini E, Bolis M, Gianni’ M, Paroni G, Fratelli M, Terao M (2016) Lipid-sensors, enigmatic-orphan and orphan nuclear receptors as therapeutic targets in breast-cancer. Oncotarget 7(27):42661–42682
Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123(3):725–731
Saal LH, Vallon-Christersson J, Häkkinen J, Hegardt C, Grabau D, Winter C, Brueffer C, Tang MH, Reuterswärd C, Schulz R et al (2015) The Sweden Cancerome Analysis Network-Breast (SCAN-B) Initiative: a large-scale multicenter infrastructure towards implementation of breast cancer genomic analyses in the clinical routine. Genome Med 7(1):20
Brueffer C, Vallon-Christersson J, Grabau D, Ehinger A, Häkkinen J, Hegardt C, Malina J, Chen Y, Bendahl P-O, Manjer J et al (2018) Clinical value of RNA sequencing-based classifiers for prediction of the five conventional breast cancer biomarkers: a report from the population-based multicenter Sweden Cancerome Analysis Network-Breast Initiative. JCO Precis Oncol 2:18
Network CGA (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61–70
Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M (2019) New approach for understanding genome variations in KEGG. Nucleic Acids Res 47(D1):D590–D595
Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28(1):27–30
Chen X, Li J, Gray WH, Lehmann BD, Bauer JA, Shyr Y, Pietenpol JA (2012) TNBCtype: a subtyping tool for triple-negative breast cancer. Cancer Inform. https://doi.org/10.4137/CIN.S9983
Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA (2011) Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Investig 121(7):2750–2767
Bailey TL (2011) DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27(12):1653–1659
Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS (2007) Quantifying similarity between motifs. Genome Biol 8(2):R24
Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Chèneby J, Kulkarni SR, Tan G et al (2018) JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res 46(D1):D260–D266
Sugita B, Gill M, Mahajan A, Duttargi A, Kirolikar S, Almeida R, Regis K, Oluwasanmi OL, Marchi F, Marian C et al (2016) Differentially expressed miRNAs in triple negative breast cancer between African-American and non-Hispanic white women. Oncotarget 7(48):79274–79291
Heckler MM, Riggins RB (2015) ERRβ splice variants differentially regulate cell cycle progression. Cell Cycle 14(1):31–45
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y et al (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486(7403):346–352
Tesarova P (2012) Breast cancer in the elderly—should it be treated differently? Rep Pract Oncol Radiother 18(1):26–33
Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR et al (2015) A global reference for human genetic variation. Nature 526(7571):68–74
Collin RW, Kalay E, Tariq M, Peters T, van der Zwaag B, Venselaar H, Oostrik J, Lee K, Ahmed ZM, Caylan R et al (2008) Mutations of ESRRB encoding estrogen-related receptor beta cause autosomal-recessive nonsyndromic hearing impairment DFNB35. Am J Hum Genet 82(1):125–138
Ben Saïd M, Ayedi L, Mnejja M, Hakim B, Khalfallah A, Charfeddine I, Khifagi C, Turki K, Ayadi H, Benzina Z et al (2011) A novel missense mutation in the ESRRB gene causes DFNB35 hearing loss in a Tunisian family. Eur J Med Genet 54(6):e535–e541
Chen F, Zhang Q, McDonald T, Davidoff MJ, Bailey W, Bai C, Liu Q, Caskey CT (1999) Identification of two hERR2-related novel nuclear receptors utilizing bioinformatics and inverse PCR. Gene 228(1–2):101–109
Zhou W, Liu Z, Wu J, Liu JH, Hyder SM, Antoniou E, Lubahn DB (2006) Identification and characterization of two novel splicing isoforms of human estrogen-related receptor beta. J Clin Endocrinol Metab 91(2):569–579
Roy B, Haupt LM, Griffiths LR (2013) Review: alternative splicing (AS) of genes as an approach for generating protein complexity. Curr Genomics 14(3):182–194
Divekar SD, Tiek DM, Fernandez A, Riggins RB (2016) Estrogen-related receptor β (ERRβ)—renaissance receptor or receptor renaissance? Nucl Recept Signal 14:e002
Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, Moses HL, Sanders ME, Pietenpol JA (2016) Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PLoS ONE 11(6):e0157368
Sem DS, Casimiro DR, Kliewer SA, Provencal J, Evans RM, Wright PE (1997) NMR spectroscopic studies of the DNA-binding domain of the monomer-binding nuclear orphan receptor, human estrogen related receptor-2 the carboxyl-terminal extension to the zinc-finger region is unstructured in the free form of the protein. J Biol Chem 272(29):18038–18043
Gearhart MD, Holmbeck SM, Evans RM, Dyson HJ, Wright PE (2003) Monomeric complex of human orphan estrogen related receptor-2 with DNA: a pseudo-dimer interface mediates extended half-site recognition. J Mol Biol 327(4):819–832
Castet A, Herledan A, Bonnet S, Jalaguier S, Vanacker J-M, Cavaillès V (2006) Receptor-interacting protein 140 differentially regulates estrogen receptor-related receptor transactivation depending on target genes. Mol Endocrinol 20(5):1035–1047
Data NRCUPoDCoRaE: improving racial and ethnic data on health: report of a workshop (2003). In: Melnick D, Perrin E (eds) Improving racial and ethnic data on health. National Academies Press, Washington, DC
Donna DY, Forman BM (2005) Identification of an agonist ligand for estrogen-related receptors ERRβ/γ. Bioorg Med Chem Lett 15(5):1311–1313
Ajay SS, Parker SC, Abaan HO, Fajardo KV, Margulies EH (2011) Accurate and comprehensive sequencing of personal genomes. Genome Res 21(9):1498–1505
Mirebrahim H, Close TJ, Lonardi S (2015) De novo meta-assembly of ultra-deep sequencing data. Bioinformatics 31(12):i9–i16
Acknowledgements
We are grateful to Allison Fitzgerald, Dr. Hillary Stires, Ayodeji Olukoya, and Sonali Persaud for their insights and/or critical reading of the manuscript. We would like to thank Drs. Bassem Haddad, Filipa Lynce, and Michael Johnson for their guidance in developing the HTSR’s invasive ductal carcinoma breast cancer tissue microarray series. We thank Henry Cho and Gaelle Palmer for their contributions to the TMA-associated REDCap database. Thank you to the Survey, Recruitment, and Biospecimen collection Shared Resource (SRBSR) for their support of research recruitment at MedStar Georgetown University Hospital. Thank you to Dr. Lao Saal of Lund University, Sweden for kindly providing the clinical information of SCAN-B data. We thank Garrett Graham for his guidance on all computational studies and Dr. Max Kushner for his aid with the DREME analysis. The results shown here are based in part upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Funding
These studies were supported in part by Department of Defense Breast Cancer Research Program Award W81XWH-17-1-0615 to RBR. Georgetown University Medical Center Shared Resources are supported in part by P30 CA051008 (Lombardi Comprehensive Cancer Center Support Grant; Principal Investigator Dr. Louis Weiner). Fellowship funding for AIF was provided by the LCCC’s Graduate Training in Breast Cancer Health Disparities Research Grant from Susan G. Komen for the Cure (GTDR15330383; Principal Investigator Lucile L. Adams-Campbell).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10549_2019_5485_MOESM1_ESM.pdf
Supplementary material 1 (PDF 4240 kb) Supplemental Fig. 1. Age at diagnosis and association of ESRRB with OS in SCAN-B and TCGA datasets. a, b Average age of patients in SCAN-B dataset by Pam50 (a) and IHC (b) subtypes. ANOVA with multiple comparisons ***p < 0.001, ****p < 0.0001. c–h KM plots showing overall survival of all breast cancer patients (c, f), BLBC patients (d, g), and TNBC patients (e, h) in SCAN-B (c–e) and TCGA (f–h) data Supplemental Fig. 2. Demographics of aCGH cohort. a–e Distribution of a age, b tumor size and ESRRB copy number, c pathology, d lymph node status and e metastasis Supplemental Fig. 3. Overlap of BLBC patients and TNBC patients. a, b Heat map representing differential expression of overlapping genes in SCAN-B versus TCGA datasets. c, d Venn diagram of patients in SCAN-B (c) and TCGA (d) data. e List of overrepresented motifs in promoter region of DEGs in SCAN-B datasets Supplemental Fig. 4. Protein and mRNA levels of ERRβ/ESRRB. Densitometry quantifying ERRβ splice variants ERRβ2 (a) and ERRβsf (b) protein levels in cell lines. cESRRB mRNA levels in SCAN-B patients, sorted into TNBC subtypes. d, eESRRB mRNA levels in cell lines representing the TNBC cell lines Supplemental Fig. 5. IHC optimization and localization. a Negative and positive controls used for antibody optimization of ERRβsf ERRβ2. Scale bar = 100 μm. b High magnification view representing subcellular staining of tumor cores staining for ERRβsf or ERRβ2. Selected panels for ERRβsf from tumor cores identified by Vectra3 as > 90% nuclear and cytoplasmic staining, or > 90% cytoplasmic staining. Arrows in left panels show positive nuclei Supplemental Table 1. DEGs in ESRRB high and low patients. List of DEGs found in BLBC and TNBC patients from SCAN-B and TCGA datasets Supplemental Table 2. ERRβ isoform expression and subcellular localization. Mean (sd) and median (IQR) expression of ERRβ2 receptor, ERRβsf receptor, and total ERRβ2:total ERRβsf expression (S2.1) and nuclear/cytoplasmic localization of ERRβ2 receptor and ERRβsf (S2.2) in three IHC breast cancer subtypes Supplemental Table 3. ERRβ isoform expression and clinical features. Analysis of ERRβ2 and ERRβsf receptor expression in the IHC subtypes and lymph node status, race, and age, with and without interaction
Rights and permissions
About this article
Cite this article
Fernandez, A.I., Geng, X., Chaldekas, K. et al. The orphan nuclear receptor estrogen-related receptor beta (ERRβ) in triple-negative breast cancer. Breast Cancer Res Treat 179, 585–604 (2020). https://doi.org/10.1007/s10549-019-05485-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-019-05485-5