Abstract
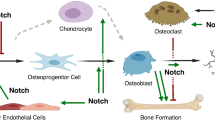
Notch (Notch1 through 4) are transmembrane receptors that play a fundamental role in cell differentiation and function. Notch receptors are activated following interactions with their ligands in neighboring cells. There are five classic ligands termed Jagged (Jag)1 and Jag2 and Delta-like (Dll)1, Dll3, and Dll4. Recent work has established Notch as a signaling pathway that plays a critical role in the differentiation and function of cells of the osteoblast and osteoclast lineages and in skeletal development and bone remodeling. The effects of Notch are cell-context dependent, and the four Notch receptors carry out specific functions in the skeleton. Gain- and loss-of-function mutations of components of the Notch signaling pathway result in a variety of congenital disorders with significant craniofacial and skeletal manifestations. The Notch ligand Jag1 is a determinant of bone mineral density, and Notch plays a role in the early phases of fracture healing. Alterations in Notch signaling are associated with osteosarcoma and with the metastatic potential of carcinoma of the breast and of the prostate. Controlling Notch signaling could prove useful in diseases of Notch gain-of-function and in selected skeletal disorders. However, clinical data on agents that modify Notch signaling are not available. In conclusion, Notch signaling is a novel pathway that regulates skeletal homeostasis in health and disease.

Similar content being viewed by others
Change history
19 October 2018
Table 2 of the original article was incorrect. The correct table is shown below.
Abbreviations
- ANK :
-
ankyrin
- BMD :
-
bone mineral density
- CHSY :
-
chondroitin sulfate synthase
- DII :
-
Delta-like
- Dkk1 :
-
Dickkopf 1
- EGF :
-
epidermal growth factor
- HES :
-
hairy and enhancer of split
- EOGT :
-
EGF-domain-specific O-linked N-acetylglucosamine transferase
- HCS :
-
Hajdu-Cheney syndrome
- HEY :
-
HES with a YRPW motif
- HD :
-
heterodimerization domain
- Jag :
-
jagged
- MAML :
-
mastermind-like
- LP :
-
leader peptide
- LMS :
-
lateral meningocele syndrome
- LNR :
-
Lin12-Notch repeats
- Lnfg :
-
lunatic fringe
- NRR :
-
negative regulatory region
- NICD :
-
Notch intracellular domain
- Nf :
-
nuclear factor
- NLS :
-
nuclear localization sequence
- PTH :
-
parathyroid hormone
- PEST :
-
proline (P)-, glutamic acid (E)-, serine (S)-, and threonine (T)-rich
- RANKL :
-
receptor activator of NF-κB ligand
- RBPJκ :
-
recombination signal-binding protein for Ig of κ region
- RAM :
-
RBPJκ-association module
- TGF :
-
transforming growth factor
- TMD :
-
transmembrane domain
References
Bai S, Kopan R, Zou W, Hilton MJ, Ong CT, Long F, Ross FP, Teitelbaum SL (2008) NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J Biol Chem 283:6509–6518
Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM, Chen Y, Wang L, Zheng H, Sutton RE, Boyce BF, Lee B (2008) Dimorphic effects of Notch signaling in bone homeostasis. Nat Med 14:299–305
Fukushima H, Nakao A, Okamoto F, Shin M, Kajiya H, Sakano S, Bigas A, Jimi E, Okabe K (2008) The association of Notch2 and NF-kappaB accelerates RANKL-induced osteoclastogenesis. Mol Cell Biol 28:6402–6412
Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, Long F (2008) Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med 14:306–314
Zanotti S, Canalis E (2016) Notch signaling and the skeleton. Endocr Rev 37:223–253
Canalis E, Adams DJ, Boskey A, Parker K, Kranz L, Zanotti S (2013) Notch signaling in osteocytes differentially regulates cancellous and cortical bone remodeling. J Biol Chem 288:25614–25625
Canalis E, Parker K, Feng JQ, Zanotti S (2013) Osteoblast lineage-specific effects of Notch activation in the skeleton. Endocrinology 154:623–634
Sanchez-Irizarry C, Carpenter AC, Weng AP, Pear WS, Aster JC, Blacklow SC (2004) Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol Cell Biol 24:9265–9273
Kopan R, Ilagan MX (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137:216–233
Siebel C, Lendahl U (2017) Notch signaling in development, tissue homeostasis, and disease. Physiol Rev 97:1235–1294
Iso T, Kedes L, Hamamori Y (2003) HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol 194:237–255
Kovall RA (2007) Structures of CSL, Notch and Mastermind proteins: piecing together an active transcription complex. Curr Opin Struct Biol 17:117–127
Wu J, Bresnick EH (2007) Bare rudiments of notch signaling: how receptor levels are regulated. Trends Biochem Sci 32:477–485
Yuan Z, Friedmann DR, Vanderwielen BD, Collins KJ, Kovall RA (2012) Characterization of CSL (CBF-1, Su(H), Lag-1) mutants reveals differences in signaling mediated by Notch1 and Notch2. J Biol Chem 287:34904–34916
Zanotti S, Canalis E (2017) Parathyroid hormone inhibits Notch signaling in osteoblasts and osteocytes. Bone 103:159–167
Deregowski V, Gazzerro E, Priest L, Rydziel S, Canalis E (2006) Notch 1 overexpression inhibits osteoblastogenesis by suppressing Wnt/beta-catenin but not bone morphogenetic protein signaling. J Biol Chem 281:6203–6210
Zanotti S, Canalis E (2014) Notch1 and Notch2 expression in osteoblast precursors regulates femoral microarchitecture. Bone 62:22–28
Canalis E, Bridgewater D, Schilling L, Zanotti S (2015) Canonical Notch activation in osteocytes causes osteopetrosis. Am J Phys Endocrinol Metab 310:E171–E182
Tu X, Delgado-Calle J, Condon KW, Maycas M, Zhang H, Carlesso N, Taketo MM, Burr DB, Plotkin LI, Bellido T (2015) Osteocytes mediate the anabolic actions of canonical Wnt/beta-catenin signaling in bone. Proc Natl Acad Sci U S A 112:E478–E486
Robling AG, Niziolek PJ, Baldridge LA, Condon KW, Allen MR, Alam I, Mantila SM, Gluhak-Heinrich J, Bellido TM, Harris SE, Turner CH (2008) Mechanical stimulation of bone in vivo reduces osteocyte expression of Sost/sclerostin. J Biol Chem 283:5866–5875
Yamada T, Yamazaki H, Yamane T, Yoshino M, Okuyama H, Tsuneto M, Kurino T, Hayashi S, Sakano S (2003) Regulation of osteoclast development by Notch signaling directed to osteoclast precursors and through stromal cells. Blood 101:2227–2234
Zhao B, Grimes SN, Li S, Hu X, Ivashkiv LB (2012) TNF-induced osteoclastogenesis and inflammatory bone resorption are inhibited by transcription factor RBP-J. J Exp Med 209:319–334
Canalis E, Schilling L, Yee SP, Lee SK, Zanotti S (2016) Hajdu Cheney mouse mutants exhibit osteopenia, increased osteoclastogenesis and bone resorption. J Biol Chem 291:1538–1551
Canalis E, Zanotti S (2017) Hairy and enhancer of split-related with YRPW motif-like (HeyL) is dispensable for bone remodeling in mice. J Cell Biochem 118:1819–1826
Salie R, Kneissel M, Vukevic M, Zamurovic N, Kramer I, Evans G, Gerwin N, Mueller M, Kinzel B, Susa M (2010) Ubiquitous overexpression of Hey1 transcription factor leads to osteopenia and chondrocyte hypertrophy in bone. Bone 46:680–694
Zanotti S, Canalis E (2013) Hairy and enhancer of split-related with YRPW motif (HEY)2 regulates bone remodeling in mice. J Biol Chem 288:21547–21557
Zanotti S, Smerdel-Ramoya A, Canalis E (2011) Hairy and enhancer of split (HES)1 is a determinant of bone mass. J Biol Chem 286:2648–2657
Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, Milner LA, Kronenberg HM, Scadden DT (2003) Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425:841–846
Duggan SP, McCarthy JV (2016) Beyond gamma-secretase activity: the multifunctional nature of presenilins in cell signalling pathways. Cell Signal 28:1–11
del Alamo D, Rouault H, Schweisguth F (2011) Mechanism and significance of cis-inhibition in Notch signalling. Curr Biol 21:R40–R47
Zanotti S, Yu J, Adhikari S, Canalis E (2018) Glucocorticoids inhibit notch target gene expression in osteoblasts. J Cell Biochem 119:6016–6023
Canalis E (2018) Clinical and experimental aspects of notch receptor signaling: Hajdu-Cheney syndrome and related disorders. Metabolism 80:48–56
Isidor B, Lindenbaum P, Pichon O, Bézieau S, Dina C, Jacquemont S, Martin-Coignard D, Thauvin-Robinet C, le Merrer M, Mandel JL, David A, Faivre L, Cormier-Daire V, Redon R, le Caignec C (2011) Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis. Nat Genet 43:306–308
Simpson MA, Irving MD, Asilmaz E, Gray MJ, Dafou D, Elmslie FV, Mansour S, Holder SE, Brain CE, Burton BK, Kim KH, Pauli RM, Aftimos S, Stewart H, Kim CA, Holder-Espinasse M, Robertson SP, Drake WM, Trembath RC (2011) Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat Genet 43:303–305
Zhao W, Petit E, Gafni RI, Collins MT, Robey PG, Seton M, Miller KK, Mannstadt M (2013) Mutations in NOTCH2 in patients with Hajdu-Cheney syndrome. Osteoporos Int 24:2275–2281
Lehman RA, Stears JC, Wesenberg RL, Nusbaum ED (1977) Familial osteosclerosis with abnormalities of the nervous system and meninges. J Pediatr 90:49–54
Gripp KW, Robbins KM, Sobreira NL et al (2015) Truncating mutations in the last exon of NOTCH3 cause lateral meningocele syndrome. Am J Med Genet A 167A:271–281
Temtamy SA, Aglan MS (2008) Brachydactyly. Orphanet J Rare Dis 3:15
Tian J, Ling L, Shboul M, Lee H, O'Connor B, Merriman B, Nelson SF, Cool S, Ababneh OH, al-Hadidy A, Masri A, Hamamy H, Reversade B (2010) Loss of CHSY1, a secreted FRINGE enzyme, causes syndromic brachydactyly in humans via increased NOTCH signaling. Am J Hum Genet 87:768–778
Stittrich AB, Lehman A, Bodian DL, Ashworth J, Zong Z, Li H, Lam P, Khromykh A, Iyer RK, Vockley JG, Baveja R, Silva ES, Dixon J, Leon EL, Solomon BD, Glusman G, Niederhuber JE, Roach JC, Patel MS (2014) Mutations in NOTCH1 cause Adams-Oliver syndrome. Am J Hum Genet 95:275–284
Hassed SJ, Wiley GB, Wang S, Lee JY, Li S, Xu W, Zhao ZJ, Mulvihill JJ, Robertson J, Warner J, Gaffney PM (2012) RBPJ mutations identified in two families affected by Adams-Oliver syndrome. Am J Hum Genet 91:391–395
Meester JA, Southgate L, Stittrich AB et al (2015) Heterozygous loss-of-function mutations in DLL4 cause Adams-Oliver syndrome. Am J Hum Genet 97:475–482
Shaheen R, Aglan M, Keppler-Noreuil K, Faqeih E, Ansari S, Horton K, Ashour A, Zaki MS, al-Zahrani F, Cueto-González AM, Abdel-Salam G, Temtamy S, Alkuraya FS (2013) Mutations in EOGT confirm the genetic heterogeneity of autosomal-recessive Adams-Oliver syndrome. Am J Hum Genet 92:598–604
Emerick KM, Rand EB, Goldmuntz E, Krantz ID, Spinner NB, Piccoli DA (1999) Features of Alagille syndrome in 92 patients: frequency and relation to prognosis. Hepatology 29:822–829
Crosnier C, Driancourt C, Raynaud N, Dhorne-Pollet S, Pollet N, Bernard O, Hadchouel M, Meunier-Rotival M (1999) Mutations in JAGGED1 gene are predominantly sporadic in Alagille syndrome. Gastroenterology 116:1141–1148
Kamath BM, Bauer RC, Loomes KM, Chao G, Gerfen J, Hutchinson A, Hardikar W, Hirschfield G, Jara P, Krantz ID, Lapunzina P, Leonard L, Ling S, Ng VL, Hoang PL, Piccoli DA, Spinner NB (2012) NOTCH2 mutations in Alagille syndrome. J Med Genet 49:138–144
McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB (2006) NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet 79:169–173
Dunwoodie SL, Clements M, Sparrow DB, Sa X, Conlon RA, Beddington RS (2002) Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development 129:1795–1806
Cornier AS, Staehling-Hampton K, Delventhal KM, Saga Y, Caubet JF, Sasaki N, Ellard S, Young E, Ramirez N, Carlo SE, Torres J, Emans JB, Turnpenny PD, Pourquié O (2008) Mutations in the MESP2 gene cause spondylothoracic dysostosis/Jarcho-Levin syndrome. Am J Hum Genet 82:1334–1341
Sparrow DB, Chapman G, Wouters MA, Whittock NV, Ellard S, Fatkin D, Turnpenny PD, Kusumi K, Sillence D, Dunwoodie SL (2006) Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am J Hum Genet 78:28–37
Sparrow DB, Guillen-Navarro E, Fatkin D, Dunwoodie SL (2008) Mutation of hairy-and-enhancer-of-Split-7 in humans causes spondylocostal dysostosis. Hum Mol Genet 17:3761–3766
Descartes M, Rojnueangnit K, Cole L, Sutton A, Morgan SL, Patry L, Samuels ME (2014) Hajdu-Cheney syndrome: phenotypical progression with de-novo NOTCH2 mutation. Clin Dysmorphol 23:88–94
Hajdu N, Kauntze R (1948) Cranio-skeletal dysplasia. Br J Radiol 21:42–48
Sargin G, Cildag S, Senturk T (2013) Hajdu-Cheney syndrome with ventricular septal defect. Kaohsiung J Med Sci 29:343–344
Sakka S, Gafni RI, Davies JH, Clarke B, Tebben P, Samuels M, Saraff V, Klaushofer K, Fratzl-Zelman N, Roschger P, Rauch F, Högler W (2017) Bone structural characteristics and response to bisphosphonate treatment in children with Hajdu-Cheney syndrome. J Clin Endocrinol Metab 102:4163–4172
Vollersen N, Hermans-Borgmeyer I, Cornils K, Fehse B, Rolvien T, Triviai I, Jeschke A, Oheim R, Amling M, Schinke T, Yorgan TA (2018) High bone turnover in mice carrying a pathogenic Notch2 mutation causing Hajdu-Cheney syndrome. J Bone Miner Res 33:70–83
Zanotti S, Yu J, Bridgewater D, Wolf JM, Canalis E (2018) Mice harboring a Hajdu Cheney syndrome mutation are sensitized to osteoarthritis. Bone 114:198–205
Galli-Tsinopoulou A, Kyrgios I, Giza S, Giannopoulou EM, Maggana I, Laliotis N (2012) Two-year cyclic infusion of pamidronate improves bone mass density and eliminates risk of fractures in a girl with osteoporosis due to Hajdu-Cheney syndrome. Minerva Endocrinol 37:283–289
McKiernan FE (2008) Integrated anti-remodeling and anabolic therapy for the osteoporosis of Hajdu-Cheney syndrome: 2-year follow-up. Osteoporos Int 19:379–380
Adami G, Rossini M, Gatti D, Orsolini G, Idolazzi L, Viapiana O, Scarpa A, Canalis E (2016) Hajdu Cheney syndrome; report of a novel NOTCH2 mutation and treatment with denosumab. Bone 92:150–156
Engin F, Bertin T, Ma O, Jiang MM, Wang L, Sutton RE, Donehower LA, Lee B (2009) Notch signaling contributes to the pathogenesis of human osteosarcomas. Hum Mol Genet 18:1464–1470
Tao J, Jiang MM, Jiang L, Salvo JS, Zeng HC, Dawson B, Bertin TK, Rao PH, Chen R, Donehower LA, Gannon F, Lee BH (2014) Notch activation as a driver of osteogenic sarcoma. Cancer Cell 26:390–401
Canalis E, Sanjay A, Yu J, Zanotti S (2017) An antibody to Notch2 reverses the osteopenic phenotype of Hajdu-Cheney mutant male mice. Endocrinology 158:730–742
Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, Schahin-Reed D, Dow GJ, Shelton A, Stawicki S, Watts RJ, Zhang J, Choy R, Howard P, Kadyk L, Yan M, Zha J, Callahan CA, Hymowitz SG, Siebel CW (2010) Therapeutic antibody targeting of individual Notch receptors. Nature 464:1052–1057
Kiel MJ, Velusamy T, Betz BL, Zhao L, Weigelin HG, Chiang MY, Huebner-Chan DR, Bailey NG, Yang DT, Bhagat G, Miranda RN, Bahler DW, Medeiros LJ, Lim MS, Elenitoba-Johnson KSJ (2012) Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J Exp Med 209:1553–1565
Yu J, Zanotti S, Schilling L, Schoenherr C, Economides AN, Sanjay A, Canalis E (2018) Induction of the Hajdu-Cheney syndrome mutation in CD19 B cells in mice alters B-cell allocation but not skeletal homeostasis. Am J Pathol 188:1430–1446
Yu J, Zanotti S, Walia B, Jellison E, Sanjay A, Canalis E (2018) The Hajdu Cheney mutation is a determinant of B-cell allocation of the splenic marginal zone. Am J Pathol 188:149–159
Gripp KW, Scott CI Jr, Hughes HE et al (1997) Lateral meningocele syndrome: three new patients and review of the literature. Am J Med Genet 70:229–239
Gripp KW (2011) Lateral meningocele syndrome and Hajdu-Cheney syndrome: different disorders with overlapping phenotypes. Am J Med Genet A 155A:1773–1774 author reply 1775
Snape KM, Ruddy D, Zenker M, Wuyts W, Whiteford M, Johnson D, Lam W, Trembath RC (2009) The spectra of clinical phenotypes in aplasia cutis congenita and terminal transverse limb defects. Am J Med Genet A 149A:1860–1881
Hoyme HE, Jones KL, Van Allen MI, Saunders BS, Benirschke K (1982) Vascular pathogenesis of transverse limb reduction defects. J Pediatr 101:839–843
Alagille D, Estrada A, Hadchouel M, Gautier M, Odievre M, Dommergues JP (1987) Syndromic paucity of interlobular bile ducts (Alagille syndrome or arteriohepatic dysplasia): review of 80 cases. J Pediatr 110:195–200
Morrissette JD, Colliton RP, Spinner NB (2001) Defective intracellular transport and processing of JAG1 missense mutations in Alagille syndrome. Hum Mol Genet 10:405–413
Boyer-Di Ponio J, Wright-Crosnier C, Groyer-Picard MT, Driancourt C, Beau I, Hadchouel M, Meunier-Rotival M (2007) Biological function of mutant forms of JAGGED1 proteins in Alagille syndrome: inhibitory effect on Notch signaling. Hum Mol Genet 16:2683–2692
Xue Y, Gao X, Lindsell CE, Norton CR, Chang B, Hicks C, Gendron-Maguire M, Rand EB, Weinmaster G, Gridley T (1999) Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet 8:723–730
McCright B, Gao X, Shen L, Lozier J, Lan Y, Maguire M, Herzlinger D, Weinmaster G, Jiang R, Gridley T (2001) Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development 128:491–502
Turnpenny PD, Whittock N, Duncan J, Dunwoodie S, Kusumi K, Ellard S (2003) Novel mutations in DLL3, a somitogenesis gene encoding a ligand for the Notch signalling pathway, cause a consistent pattern of abnormal vertebral segmentation in spondylocostal dysostosis. J Med Genet 40:333–339
Kusumi K, Sun ES, Kerrebrock AW, Bronson RT, Chi DC, Bulotsky MS, Spencer JB, Birren BW, Frankel WN, Lander ES (1998) The mouse pudgy mutation disrupts Delta homologue Dll3 and initiation of early somite boundaries. Nat Genet 19:274–278
Saga Y, Hata N, Koseki H, Taketo MM (1997) Mesp2: a novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes Dev 11:1827–1839
Kung AW, Xiao SM, Cherny S et al (2010) Association of JAG1 with bone mineral density and osteoporotic fractures: a genome-wide association study and follow-up replication studies. Am J Hum Genet 86:229–239
Dishowitz MI, Terkhorn SP, Bostic SA, Hankenson KD (2012) Notch signaling components are upregulated during both endochondral and intramembranous bone regeneration. J Orthop Res 30:296–303
Wang C, Shen J, Yukata K, Inzana JA, O'Keefe RJ, Awad HA, Hilton MJ (2015) Transient gamma-secretase inhibition accelerates and enhances fracture repair likely via Notch signaling modulation. Bone 73:77–89
Wang C, Inzana JA, Mirando AJ, Ren Y, Liu Z, Shen J, O'Keefe RJ, Awad HA, Hilton MJ (2016) NOTCH signaling in skeletal progenitors is critical for fracture repair. J Clin Invest 126:1471–1481
Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, Mackem S, Carmeliet G, Kronenberg HM (2010) Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell 19:329–344
Ramasamy SK, Kusumbe AP, Wang L, Adams RH (2014) Endothelial Notch activity promotes angiogenesis and osteogenesis in bone. Nature 507:376–380
Dishowitz MI, Mutyaba PL, Takacs JD, Barr AM, Engiles JB, Ahn J, Hankenson KD (2013) Systemic inhibition of canonical Notch signaling results in sustained callus inflammation and alters multiple phases of fracture healing. PLoS One 8:e68726
Tanaka M, Setoguchi T, Hirotsu M, Gao H, Sasaki H, Matsunoshita Y, Komiya S (2009) Inhibition of Notch pathway prevents osteosarcoma growth by cell cycle regulation. Br J Cancer 100:1957–1965
Dailey DD, Anfinsen KP, Pfaff LE, Ehrhart EJ, Charles JB, Bonsdorff TB, Thamm DH, Powers BE, Jonasdottir TJ, Duval DL (2013) HES1, a target of Notch signaling, is elevated in canine osteosarcoma, but reduced in the most aggressive tumors. BMC Vet Res 9:130
Hughes DP (2009) How the NOTCH pathway contributes to the ability of osteosarcoma cells to metastasize. Cancer Treat Res 152:479–496
Zhang Z, Wang H, Ikeda S, Fahey F, Bielenberg D, Smits P, Hauschka PV (2010) Notch3 in human breast cancer cell lines regulates osteoblast-cancer cell interactions and osteolytic bone metastasis. Am J Pathol 177:1459–1469
Sethi N, Dai X, Winter CG, Kang Y (2011) Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell 19:192–205
Zayzafoon M, Abdulkadir SA, McDonald JM (2004) Notch signaling and ERK activation are important for the osteomimetic properties of prostate cancer bone metastatic cell lines. J Biol Chem 279:3662–3670
Ryeom SW (2011) The cautionary tale of side effects of chronic Notch1 inhibition. J Clin Invest 121:508–509
De Strooper B, Annaert W, Cupers P et al (1999) A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398:518–522
Ilagan MX, Kopan R (2013) Selective blockade of transport via SERCA inhibition: the answer for oncogenic forms of Notch? Cancer Cell 23:267–269
Moellering RE, Cornejo M, Davis TN, Del BC, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE (2009) Direct inhibition of the NOTCH transcription factor complex. Nature 462:182–188
Li K, Li Y, Wu W, Gordon WR, Chang DW, Lu M, Scoggin S, Fu T, Vien L, Histen G, Zheng J, Martin-Hollister R, Duensing T, Singh S, Blacklow SC, Yao Z, Aster JC, Zhou BBS (2008) Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J Biol Chem 283:8046–8054
Zanotti S, Canalis E (2010) Notch and the skeleton. Mol Cell Biol 30:886–896
Aste-Amezaga M, Zhang N, Lineberger JE et al (2010) Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PLoS One 5:e9094
Yan M, Callahan CA, Beyer JC, Allamneni KP, Zhang G, Ridgway JB, Niessen K, Plowman GD (2010) Chronic DLL4 blockade induces vascular neoplasms. Nature 463:E6–E7
Acknowledgments
The author thanks Mary Yurczak for secretarial assistance.
Funding
This work was supported by Grants AR063049, AR068160, and AR072987 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases and Grant DK045227 from the National Institute of Diabetes and Digestive and Kidney Diseases.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None.
Rights and permissions
About this article
Cite this article
Canalis, E. Notch in skeletal physiology and disease. Osteoporos Int 29, 2611–2621 (2018). https://doi.org/10.1007/s00198-018-4694-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00198-018-4694-3