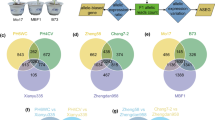
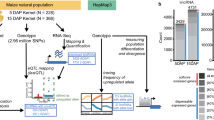
Abstract
Hybrid vigor, also known as heterosis, has been widely utilized in agronomic production of maize (Zea mays L.) and other crops. However, the molecular mechanisms underlying heterosis are still not fully understood. To provide a more complete understanding of the transcriptomic dynamics associated with heterosis, we collected a comprehensive set of sequence data on linear mRNA transcripts and circular RNAs (circRNAs) from seedling leaves of two widely used maize inbred lines and their F1 hybrid at the V4 growth stage. We detected over 25,000 expressed genes with more than 1200 circRNAs that showed dramatic and distinct variations in expression level across the three genotypes. Although most linear and circular transcripts exhibited additive expression in the hybrid, the expression of circRNAs was more likely to be nonadditive. Interestingly, the levels of linear transcripts and their corresponding circRNAs from the same loci showed a significant relationship and coordinated expression mode across all three genotypes. Notably, in the hybrid, allele-specific expression of linear transcripts was significantly associated with the expression of circRNAs from the same locus, suggesting potential regulatory cross talk between linear and circular transcripts. Our study provides a deeper understanding of dynamic variations for both the linear and circular transcriptome in a classical hybrid triplet of maize.




Similar content being viewed by others
References
Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, Gorospe M (2017) Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 14:361–369
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56:55–66
Auger DL, Gray AD, Ream TS, Kato A, Coe EH Jr, Birchler JA (2005) Nonadditive gene expression in diploid and triploid hybrids of maize. Genetics 169:389–397
Baldauf JA, Marcon C, Paschold A, Hochholdinger F (2016) Nonsyntenic genes drive tissue-specific dynamics of differential, nonadditive, and allelic expression patterns in maize hybrids. Plant Physiol 171:1144–1155
Baldauf JA, Marcon C, Lithio A, Vedder L, Altrogge L, Piepho HP, Schoof H, Nettleton D, Hochholdinger F (2018) Single-parent expression is a general mechanism driving extensive complementation of non-syntenic genes in maize hybrids. Curr Biol 28(431–437):e434
Belting HG, Shashikant CS, Ruddle FH (1998) Modification of expression and cis-regulation of Hoxc8 in the evolution of diverged axial morphology. Proc Natl Acad Sci USA 95:2355–2360
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE (2010) Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 6:e1001233
Chen LL (2016) The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17:205–211
Chen WJ, Chang SH, Hudson ME, Kwan WK, Li J, Estes B, Knoll D, Shi L, Zhu T (2005) Contribution of transcriptional regulation to natural variations in Arabidopsis. Genome Biol 6:R32
Chen I, Chen CY, Chuang TJ (2015) Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA 6:563–579
Chen L, Zhang P, Fan Y, Lu Q, Li Q, Yan J, Muehlbauer GJ, Schnable PS, Dai M, Li L (2018a) Circular RNAs mediated by transposons are associated with transcriptomic and phenotypic variation in maize. New Phytol 217:1292–1306
Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S, Song W, Li X, Li L, Du Z, Jia L, Zhou L, Li W, Hoffman AR, Hu JF, Cui J (2018b) A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol 19:218
Chu Q, Zhang X, Zhu X, Liu C, Mao L, Ye C, Zhu QH, Fan L (2017) PlantcircBase: a database for plant circular RNAs. Mol Plant 10:1126–1128
Chu Q, Bai P, Zhu X, Zhang X, Mao L, Zhu QH, Fan L, Ye CY (2018) Characteristics of plant circular RNAs. Brief Bioinform. https://doi.org/10.1093/bib/bby111
Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M (2009) Mapping complex disease traits with global gene expression. Nat Rev Genet 10:184–194
Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38:W64–W70
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB (2017) Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ 24:357–370
Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L, Lyu J, Li F, Peng C, Krylov SN, Xie Y, Zhang Y, He C, Wu N, Zhang C, Sdiri M, Dong J, Ma J, Gao C, Hibberd S, Yang BB (2018) A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene 37:5829–5842
Falconer DS (1989) Introduction to quantitative genetics, 3rd edn. Longman, England
Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289:85–88
Fu J, Cheng Y, Linghu J, Yang X, Kang L, Zhang Z, Zhang J, He C, Du X, Peng Z, Wang B, Zhai L, Dai C, Xu J, Wang W, Li X, Zheng J, Chen L, Luo L, Liu J, Qian X, Yan J, Wang J, Wang G (2013) RNA sequencing reveals the complex regulatory network in the maize kernel. Nat Commun 4:2832
Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, Ware DH, Buckler ES (2009) A first-generation haplotype map of maize. Science 326:1115–1117
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388
Hirsch CN, Foerster JM, Johnson JM, Sekhon RS, Muttoni G, Vaillancourt B, Penagaricano F, Lindquist E, Pedraza MA, Barry K, de Leon N, Kaeppler SM, Buell CR (2014) Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26:121–135
Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, Gabel G, Beutner F, Scholz M, Thiery J, Musunuru K, Krohn K, Mann M, Teupser D (2016) Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 7:12429
Holloway B, Luck S, Beatty M, Rafalski JA, Li B (2011) Genome-wide expression quantitative trait loci (eQTL) analysis in maize. BMC Genom 12:336
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ (2017) Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res 77:2339–2350
Hsu MT, Coca-Prados M (1979) Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280:339–340
Hu X, Wang H, Diao X, Liu Z, Li K, Wu Y, Liang Q, Wang H, Huang C (2016) Transcriptome profiling and comparison of maize ear heterosis during the spikelet and floret differentiation stages. BMC Genom 17:959
Huang Y, Zhang L, Zhang J, Yuan D, Xu C, Li X, Zhou D, Wang S, Zhang Q (2006) Heterosis and polymorphisms of gene expression in an elite rice hybrid as revealed by a microarray analysis of 9198 unique ESTs. Plant Mol Biol 62:579–591
Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, Mueller M, Hummel O, Monti J, Zidek V, Musilova A, Kren V, Causton H, Game L, Born G, Schmidt S, Muller A, Cook SA, Kurtz TW, Whittaker J, Pravenec M, Aitman TJ (2005) Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet 37:243–253
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–157
Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, Campbell MS, Stein JC, Wei X, Chin CS, Guill K, Regulski M, Kumari S, Olson A, Gent J, Schneider KL, Wolfgruber TK, May MR, Springer NM, Antoniou E, McCombie WR, Presting GG, McMullen M, Ross-Ibarra J, Dawe RK, Hastie A, Rank DR, Ware D (2017) Improved maize reference genome with single-molecule technologies. Nature 546:524–527
Kaeppler S (2012) Heterosis: many genes, many mechanisms—end the search for an undiscovered unifying theory. ISRN Botany 2012:1–12
Kleaveland B, Shi CY, Stefano J, Bartel DP (2018) A network of noncoding regulatory RNAs acts in the mammalian brain. Cell 174:350–362
Kristensen LS, Okholm TLH, Veno MT, Kjems J (2018) Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol 15:280–291
Lai J, Li R, Xu X, Jin W, Xu M, Zhao H, Xiang Z, Song W, Ying K, Zhang M, Jiao Y, Ni P, Zhang J, Li D, Guo X, Ye K, Jian M, Wang B, Zheng H, Liang H, Zhang X, Wang S, Chen S, Li J, Fu Y, Springer NM, Yang H, Wang J, Dai J, Schnable PS, Wang J (2010) Genome-wide patterns of genetic variation among elite maize inbred lines. Nat Genet 42:1027–1030
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I (2017) Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66(22–37):e29
Li L, Petsch K, Shimizu R, Liu S, Xu WW, Ying K, Yu J, Scanlon MJ, Schnable PS, Timmermans MC, Springer NM, Muehlbauer GJ (2013) Mendelian and non-Mendelian regulation of gene expression in maize. PLoS Genet 9:e1003202
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22:256–264
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G (2017) Corrigendum: exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 24:194
Liu Z, Ran Y, Tao C, Li S, Chen J, Yang E (2019) Detection of circular RNA expression and related quantitative trait loci in the human dorsolateral prefrontal cortex. Genome Biol 20:99
Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, Zhao Q, Zhou C, Zhao Y, Lu D, Luo J, Wang Y, Tian Q, Feng Q, Huang T, Han B (2015) Transcriptome-wide investigation of circular RNAs in rice. RNA 21:2076–2087
Luo Z, Han L, Qian J, Li L (2019) Circular RNAs exhibit extensive intraspecific variation in maize. Planta 250:69–78
Marcon C, Paschold A, Malik WA, Lithio A, Baldauf JA, Altrogge L, Opitz N, Lanz C, Schoof H, Nettleton D, Piepho HP, Hochholdinger F (2017) Stability of single-parent gene expression complementation in maize hybrids upon water deficit stress. Plant Physiol 173:1247–1257
Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18:1509–1517
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338
Meyer S, Pospisil H, Scholten S (2007) Heterosis associated gene expression in maize embryos 6 days after fertilization exhibits additive, dominant and overdominant pattern. Plant Mol Biol 63:381–391
Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S (2017) Translation of circRNAs. Mol Cell 66(9–21):e27
Paschold A, Jia Y, Marcon C, Lund S, Larson NB, Yeh CT, Ossowski S, Lanz C, Nettleton D, Schnable PS, Hochholdinger F (2012) Complementation contributes to transcriptome complexity in maize (Zea mays L.) hybrids relative to their inbred parents. Genome Res 22:2445–2454
Paschold A, Larson NB, Marcon C, Schnable JC, Yeh CT, Lanz C, Nettleton D, Piepho HP, Schnable PS, Hochholdinger F (2014) Nonsyntenic genes drive highly dynamic complementation of gene expression in maize hybrids. Plant Cell 26:3939–3948
Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, Trimbuch T, Zywitza V, Plass M, Schreyer L, Ayoub S, Kocks C, Kuhn R, Rosenmund C, Birchmeier C, Rajewsky N (2017) Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357:eaam8526
Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, Linsley PS, Mao M, Stoughton RB, Friend SH (2003) Genetics of gene expression surveyed in maize, mouse and man. Nature 422:297–302
Schnable PS, Springer NM (2013) Progress toward understanding heterosis in crop plants. Annu Rev Plant Biol 64:71–88
Schnable JL, Zhikai (2016) Maize Hybrid Imaging. figshare
Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ (2005) Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol 6:10
Shao L, Xing F, Xu C, Zhang Q, Che J, Wang X, Song J, Li X, Xiao J, Chen LL, Ouyang Y, Zhang Q (2019) Patterns of genome-wide allele-specific expression in hybrid rice and the implications on the genetic basis of heterosis. Proc Natl Acad Sci USA 116:5653–5658
Song R, Messing J (2003) Gene expression of a gene family in maize based on noncollinear haplotypes. Proc Natl Acad Sci USA 100:9055–9060
Springer NM, Stupar RM (2007) Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell 19:2391–2402
Stoll L, Sobel J, Rodriguez-Trejo A, Guay C, Lee K, Veno MT, Kjems J, Laybutt DR, Regazzi R (2018) Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol Metab 9:69–83
Stupar RM, Springer NM (2006) Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173:2199–2210
Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS (2006) All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci USA 103:6805–6810
Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucl Acids Res 45:W122–W129
Uzarowska A, Keller B, Piepho HP, Schwarz G, Ingvardsen C, Wenzel G, Lubberstedt T (2007) Comparative expression profiling in meristems of inbred-hybrid triplets of maize based on morphological investigations of heterosis for plant height. Plant Mol Biol 63:21–34
Verduci L, Ferraiuolo M, Sacconi A, Ganci F, Vitale J, Colombo T, Paci P, Strano S, Macino G, Rajewsky N, Blandino G (2017) The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD transcription-competent complex. Genome Biol 18:237
Vuylsteke M, van Eeuwijk F, Van Hummelen P, Kuiper M, Zabeau M (2005) Genetic analysis of variation in gene expression in Arabidopsis thaliana. Genetics 171:1267–1275
Wang RL, Stec A, Hey J, Lukens L, Doebley J (1999) The limits of selection during maize domestication. Nature 398:236–239
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Wang J, Yu H, Xie W, Xing Y, Yu S, Xu C, Li X, Xiao J, Zhang Q (2010) A global analysis of QTLs for expression variations in rice shoots at the early seedling stage. Plant J 63:1063–1074
Waters AJ, Bilinski P, Eichten SR, Vaughn MW, Ross-Ibarra J, Gehring M, Springer NM (2013) Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species. Proc Natl Acad Sci U S A 110:19639–19644
Waters AJ, Makarevitch I, Noshay J, Burghardt LT, Hirsch CN, Hirsch CD, Springer NM (2017) Natural variation for gene expression responses to abiotic stress in maize. Plant J 89:706–717
Wei L, Cao X (2016) The effect of transposable elements on phenotypic variation: insights from plants to humans. Sci China Life Sci 59:24–37
Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y, Goel A (2017) Circular RNA ciRS-7—a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res 23:3918–3928
Wittkopp PJ, Haerum BK, Clark AG (2004) Evolutionary changes in cis and trans gene regulation. Nature 430:85–88
Xia P, Wang S, Ye B, Du Y, Li C, Xiong Z, Qu Y, Fan Z (2018) A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity 48(688–701):e687
Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, Yang H, Wang T, Lee AY, Swanson SA, Zhang J, Zhu Y, Kim A, Nery JR, Urich MA, Kuan S, Yen CA, Klugman S, Yu P, Suknuntha K, Propson NE, Chen H, Edsall LE, Wagner U, Li Y, Ye Z, Kulkarni A, Xuan Z, Chung WY, Chi NC, Antosiewicz-Bourget JE, Slukvin I, Stewart R, Zhang MQ, Wang W, Thomson JA, Ecker JR, Ren B (2013) Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153:1134–1148
Xu H, Guo S, Li W, Yu P (2015) The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 5:12453
Yang H, Wang X, Wei Y, Deng Z, Liu H, Chen J, Dai L, Xia Z, He G, Li D (2018a) Transcriptomic analyses reveal molecular mechanisms underlying growth heterosis and weakness of rubber tree seedlings. BMC Plant Biol 18:10
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, Huang S, Xie B, Zhang N (2018b) Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst 110:304–315
Ye CY, Chen L, Liu C, Zhu QH, Fan L (2015) Widespread noncoding circular RNAs in plants. New Phytol 208:88–95
Yu CY, Li TC, Wu YY, Yeh CH, Chiang W, Chuang CY, Kuo HC (2017) The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun 8:1149
Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, Yang W, Zhang C, Yang Q, Yee A, Chen Y, Yang F, Sun H, Huang R, Yee AJ, Li RK, Wu Z, Backx PH, Yang BB (2017) A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics 7:3842–3855
Zhang TF, Li B, Zhang DF, Jia GQ, Li ZY, Wang SC (2012) Genome-wide transcriptional analysis of yield and heterosis-associated genes in maize (Zea mays L.). J Integr Agric 11:1245–1256
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL (2013) Circular intronic long noncoding RNAs. Mol Cell 51:792–806
Zhang C, Lin C, Fu F, Zhong X, Peng B, Yan H, Zhang J, Zhang W, Wang P, Ding X, Zhang W, Zhao L (2017) Comparative transcriptome analysis of flower heterosis in two soybean F1 hybrids by RNA-seq. PLoS ONE 12:e0181061
Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, Li Z, Ming L, Xie B, Zhang N (2018a) A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37:1805–1814
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, Liu H, Xu J, Xiao F, Zhou H, Yang X, Huang N, Liu J, He K, Xie K, Zhang G, Huang S, Zhang N (2018b) A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun 9:4475
Zhang P, Fan Y, Sun X, Chen L, Terzaghi W, Bucher E, Li L, Dai M (2019) A large-scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis. Plant J 98:697–713
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7:11215
Zhou P, Hirsch CN, Briggs SP, Springer NM (2019) Dynamic patterns of gene expression additivity and regulatory variation throughout maize development. Mol Plant 12:410–425
Acknowledgements
This research was supported by the National Key Research and Development Program of China (2016YFD0100800), the National Natural Science Foundation of China (31771798), the Fundamental Research Funds for the Central Universities (2662016PY096), and Huazhong Agricultural University Scientific & Technological Self-Innovation Foundation (2015RC016).We are grateful to all the partners who shared their materials and the original scientists who generated these widely used inbreds.
Author information
Authors and Affiliations
Contributions
LL designed and supervised this study. ZL and JQ collected all the data. ZL, JQ, and SC performed the data analysis. LL and ZL prepared the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Mingliang Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Luo, Z., Qian, J., Chen, S. et al. Dynamic patterns of circular and linear RNAs in maize hybrid and parental lines. Theor Appl Genet 133, 593–604 (2020). https://doi.org/10.1007/s00122-019-03489-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03489-9