Abstract
3-Hydroxy-3-methylglutaryl-coenzymeA reductase (HMGR), the first rate-limiting enzyme of Mevalonate (MVA) pathway was isolated from Andrographis paniculata (ApHMGR) and expressed in bacterial cells. Full length ApHMGR (1937 bp) was submitted to NCBI with accession number MG271748.1. The open reading frame (ORF) was flanked by a 31-bp 5′-UTR, 118-bp 3′-UTR and ApHMGR contained a 1787 bp ORF encoding protein of 595 amino acids. ApHMGR protein was approximately 64 kDa, with isoelectric point of 5.75. Isolated ApHMGR was cloned into pET102 vector and expressed in E. coli BL21 (DE 3) cells, and characterized by SDS-PAGE. HPLC analysis for andrographolide content in leaf, stem and root of A. paniculata revealed highest in leaf tissue. The expression patterns of ApHMGR in different plant tissues using qRT-PCR revealed high in root tissue correlating with HPLC data. Three dimensional (3D) structural model of ApHMGR displayed 90% of the amino acids in most favored regions of the Ramachandran plot with 93% overall quality factor. ApHMGR was highly conserved with plant specific N-terminal membrane domains and C-terminal catalytic regions. Phylogenetic analysis showed A. paniculata sharing common ancestor with Handroanthus impetiginosus. 3D model of ApHMGR was screened for the interaction with substrates NADPH, HMG CoA and inhibitor using Auto Dock Vina. In silico analysis revealed that full length ApHMGR had extensive similarities to other plant HMGRs. The present communication reports the isolation of full length HMGR from A. paniculata, its heterologous expression in bacterial cells and in silico structural and functional characterization providing valuable genomic information for future molecular interventions.

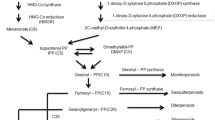
(Pathway source: Srivatsava and Akhila 2010). (Color figure online)















Similar content being viewed by others
References
Singh A, Meena AK, Sudeep Meena, Pant P, Padhi MM (2012) Studies on standardization of Andrographis paniculata nees and identification by HPTLC using andrographolide as marker compound. Int J Phram Pharm Sci. 4:197–200
Akbar S (2012) Andrographis paniculata: a review of pharmacological activities and clinical e!ects. Altern Med Rev 16:66–77
Subramanian R, Asmawi MZ, Sadikun A (2012) A bitter plant with a sweet future? A comprehensive review of an oriental medicinal plant: Andrographis paniculata. Phytochem Rev 11:39–75
Chao WW, Lin BF (2010) Isolation and identification of bioactive compounds in Andrographis paniculata (Chuanxinlian). Chin Med 5:1–15
Gupta S, Mishra KP, Ganju L (2017) Broad-spectrum antiviral properties of andrographolide. Arch Virol 162:611–623
Nitave SA, Nilesh B, Chougule Koumaravelou (2018) Phytochemical investigation, analgesic and antipyretic activities of ethanolic extract of kariyat. Int J Pharm Sci Res 9:1035–1043
Suresh V, Raju D, Karuna R et al (2016) Evaluation of antitumor potential of forskolin and andrographolide employing potato tumor bioassay model. Clin Cancer Drugs 3:121–130
Doi H, Matsui T, Ohye T, Imamura S et al (2018) Andrographolide from Andrographis paniculata shows anti-proliferative activity and anti-inflammatory effects in human monocytic leukemia cells. Fujita Med J 4:55–60
Forestier-Román IS, López-Rivas A, Sánchez-Vázquez MM et al (2019) Andrographolide induces DNA damage in prostate cancer cells. Oncotarget 10:1085–1101
Chassagne F, Haddad M, Amiel A et al (2018) A metabolomic approach to identify anti-hepatocarcinogenic compounds from plants used traditionally in the treatment of liver diseases. Fitoterapia 127:226–236
Khan I, Yusuf MA, Ansari IA, Akhtar MS (2018) Potential of andrographolide, a diterpenoid lactone from Andrographis paniculata: a nature’s treasure for chemoprevention and therapeutics. In: Akhtar MS, Swamy MK (eds) Anticancer plants: mechanisms and molecular interactions, Vol 4. Springer, pp 143–164
Singh S, Pandey P, Ghosh S, Banerjee S (2018) Anti-cancer labdane diterpenoids from adventitious roots of Andrographis paniculata: augmentation of production prospect endowed with pathway gene expression. Protoplasma 255:1387–1400
Rahman UM, Ayoob I, Rehman US et al (2018) Microwave-assisted synthesis of andrographolide analogues as potent β-Glycosidase inhibitors. SynOpen 2:200–206
Gao Z, Yu C, Liang H et al (2018) Andrographolide derivative CX-10 ameliorates dextran sulphate sodium-induced ulcerative colitis in mice: involvement of NF-κB and MAPK signalling pathways. Int Immunopharmacol 57:82–90
Liu Y, Liang RM, Ma QP et al (2017) Synthesis of thioether andrographolide derivatives and their inhibitory effect against cancer cells. Med Chem Comm 8:1268–1274
Kandanur SG, Kundu S, Cadena C, San Juan H, Bajaj A, Guzman JD, Nanduri S, Golakoti NR (2019) Design, synthesis, and biological evaluation of new 12-substituted-14-deoxy-andrographolide derivatives as apoptosis inducers. Chem Pap 73:1669–1675
Lee AR, Kwon M, Kang MK, Kim J, Kim SU, Ro DK (2019) Increased sesqui-and triterpene production by co-expression of HMG-CoA reductase and biotin carboxyl carrier protein in tobacco (Nicotiana benthamiana). Metab Eng 52:20–28
Yin JL, Wong WS (2019) Production of santalenes and bergamotene in Nicotiana tabacum plants. PLoS ONE 14:e0203249
Bansal S, Narnoliya LK, Mishra B et al (2018) HMG-CoA reductase from Camphor Tulsi (Ocimum kilimandscharicum) regulated MVA dependent biosynthesis of diverse terpenoids in homologous and heterologous plant systems. Sci Rep 8:1–15
Jayashree R, Nazeem PA, Rekha K et al (2018) Over-expression of 3-hydroxy-3- methylglutaryl-coenzyme A reductase 1 (hmgr1) gene under super-promoter for enhanced latex biosynthesis in rubber tree (Hevea brasiliensis Muell. Arg.). Plant Physiol Biochem 127:414–424
Yang R, Yuan B, Li W et al (2018) Improving the accumulation of 18 α -and 18 β -glycyrrhizins by over-expressing GuHMGR, GuSQS 1, and GuBAS genes in Glycyrrhiza uralensis. J Tradit Chin Med Sci 4:336–349
Mahobia A, Jha Z (2018) Docking studies of andrographolide and its in vitro validation. Int J Chem Stud 6:1164–1166
Sharma E, Pandey S, Gaur AK (2016) In silico characterization and differential expression pattern analysis of conserved HMG CoA reductase domain isolated from Aconitum balfourii Stapf. 3 Biotech 6:1–8
Bindu BB, Srinath M, Shailaja A, Giri CC (2017) Comparative protein profile studies and in silico structural/functional analysis of HMGR (ApHMGR) in Andrographis paniculata (Burm. f.) Wall. ex Nees. Ann Phytomed 6:30–44
Srinath M, Shailaja A, Bindu BB, Giri CC (2017) Characterization of 1-deoxy-D-xylulose 5-phosphate synthase (DXS) protein in Andrographis paniculata (Burm. f.) Wall. ex. Nees: A in silico appraisal. Ann Phytomed Int J 6:63–73
Shailaja A, Bindu BB, Srinath M, Giri CC (2018) In silico structural and functional analysis of copalyl diphosphate synthase enzyme in Andrographis paniculata (Burm. f.) Wall. ex Nees: a plant of immense pharmaceutical value. Ann Phytomed Int J 7:69–77
Cui Y, Wang Y, Ouyang X, Han Y, Zhu H, Chen Q (2009) Fingerprint profile of active components for Andrographis paniculata Nees by HPLC-DAD. Sens Instrum Food Qual Saf 3:165–179
Garg A, Agrawal L, Misra RC, Sharma S, Ghosh S (2015) Andrographis paniculata transcriptome provides molecular insights into tissue-specific accumulation of medicinal diterpenes. BMC Genomics 16:659
Mou W, Li D, Bu J, Jiang Y, Khan ZU, Luo Z, Mao L, Ying T (2016) Comprehensive analysis of ABA effects on ethylene biosynthesis and signaling during tomato fruit ripening. PLoS ONE 11:1–30
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Chojnacki S, Cowley A, Lee J, Foix A, Lopez R (2017) Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res 45:W550–W553
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer 3. Bioinformatics 23:1289–1291
Untergasser A, Cutcutache I, Koressaar T et al (2012) Primer3 - new capabilities and interfaces. Nucleic Acids Res 40:e115
San Millán RM, Martínez-Ballesteros I, Rementeria A et al (2013) Online exercise for the design and simulation of PCR and PCR-RFLP experiments. BMC Res Notes 6:513
Waterhouse A, Bertoni M, Bienert S et al (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303
Roy A, Yang J, Zhang Y (2012) COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res 40:471–477
Watson JD, Sanderson S, Ezersky A et al (2007) Towards fully automated structure-based function prediction in structural genomics: a case study. J Mol Biol 367:1511–1522
Timothy L. Bailey and Charles Elkan (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers”, Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, AAAI Press, Menlo Park, California, pp.28-36
Marchler-Bauer A, Bo Y, Han L et al (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203
Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324
Wass MN, Kelley LA, Sternberg MJ (2010) 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acid Res 38:W469–W473
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Boratyn GM, Schäffer AA, Agarwala R et al (2012) Domain enhanced lookup time accelerated BLAST. Biol Direct 7:12
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Dereeper A, Guignon V, Blanc G et al (2008) Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469
Dereeper A, Audic S, Claverie JM, Blanc G (2010) BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol 12(10):8
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408
Zhao Q, Li R, Chen X, Yang Q, Li J (2018) Cloning and characterization of the gene encoding 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase from Fritillaria cirrhosa D Don. Braz Arch Biol Technol 61:1–12
Devi K, Patar L, Modi MK, Sen P (2017) An insight into structure, sunction, and expression analysis of 3-hydroxy-3-methylglutaryl-CoA reductase of Cymbopogon winterianus. Bioinform Biol Insights. https://doi.org/10.1177/1177932217701735
Kalita R, Patar L, Shasany AK et al (2015) Molecular cloning, characterization and expression analysis of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Centella asiatica L. Mol Biol Rep 42:1431–1439
Ferrero S, Grados-Torrez RE, Leivar P et al (2015) Proliferation and morphogenesis of the endoplasmic reticulum driven by the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase in plant cells. Plant Physiol 168:899–914
Gu W, Geng C, Xue W et al (2015) Characterization and function of the 3-hydroxy-3-methylglutaryl-CoA reductase gene in Alisma orientale (Sam.) Juz. and its relationship with protostane triterpene production. Plant Physiol Biochem 97:378–389
Tiski I, Marraccini P, Pot D et al (2011) Characterization and expression of two cdna encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase isoforms in Coffee (Coffea arabica L.). Omi A J Integr Biol 15:719–727
Robertlee J, Kobayashi K, Suzuki M, Muranaka T (2017) AKIN10, a representative Arabidopsis SNF1-related protein kinase 1 (SnRK1), phosphorylates and downregulates plant HMG-CoA reductase. FEBS Lett 591:1159–1166
Srivastava N, Akhila A (2010) Biosynthesis of andrographolide in Andrographis paniculata. Phytochemistry 71:1298-304.
Zeidabadi DD, Javaran MJ, Dehghani H et al (2018) An investigation of the HMGR gene and IPI gene expression in black caraway (Bunium persicum). 3 Biotech 8:405–413
Zhang G, Wu Y, Muhammad ZU, Yang Y, Yu J, Zhang J, Yang D (2019) cDNA cloning, prokaryotic expression and functional analysis of 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR) in Pogostemon cablin. Protein Expr Purif 163:105454
Rao S, Meng X, Liao Y, Yu T, Cao J, Tan J, Xu F, Cheng S (2019) Characterization and functional analysis of two novel 3-hydroxy-3-methylglutaryl-coenzyme A reductase genes (GbHMGR2 and GbHMGR3) from Ginkgo biloba. Sci Rep 9:1–3
Acknowledgements
Authors would like to thank DST-PURSE-II sponsored by Department of Science and Technology (DST), and OU-UGC-CPEPA, UGC-BSR-RFSMS sponsored by University Grants Commission (UGC) New Delhi for financial support and fellowship to MS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. In particular, we confirm that none of the authors have any-non-financial conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Srinath, M., Bindu, B.B.V., Shailaja, A. et al. Isolation, characterization and in silico analysis of 3-Hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR) gene from Andrographis paniculata (Burm. f) Nees. Mol Biol Rep 47, 639–654 (2020). https://doi.org/10.1007/s11033-019-05172-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-05172-0