Abstract
Females and males differ substantially in various neuronal functions in divergent, sexually dimorphic animal species, including humans. Despite its developmental, physiological and medical significance, understanding the molecular mechanisms by which sex-specific differences in the anatomy and operation of the nervous system are established remains a fundamental problem in biology. Here, we show that in Caenorhabditis elegans (nematodes), the global sex-determining factor TRA-1 regulates food leaving (mate searching), male mating and adaptation to odorants in a sex-specific manner by repressing the expression of goa-1 gene, which encodes the Gα(i/o) subunit of heterotrimeric G (guanine–nucleotide binding) proteins triggering physiological responses elicited by diverse neurotransmitters and sensory stimuli. Mutations in tra-1 and goa-1 decouple behavioural patterns from the number of X chromosomes. TRA-1 binds to a conserved binding site located in the goa-1 coding region, and downregulates goa-1 expression in hermaphrodites, particularly during embryogenesis when neuronal development largely occurs. These data suggest that the sex-determination machinery is an important modulator of heterotrimeric G protein-mediated signalling and thereby various neuronal functions in this organism and perhaps in other animal phyla.
Similar content being viewed by others
Introduction
A fascinating but mechanistically largely unexplored phenomenon in neurobiology is that the two sexes differ significantly in numerous neuronal functions in sexually dimorphic animal species (Bao and Swaab 2010). Such functions include the control of behaviour, sensory information transmission, learning and memory processing. To provide human examples, women in general are less aggressive, tend to have a better sense of smell, and learn skills quicker and at an earlier age than men (Lonsdorf et al. 2004; Doty and Cameron 2009). Drug addiction and tendency to acquire certain neurodevelopmental disorders like autism are also associated with an apparent sex bias (Fattore and Fratta 2010; Zuo et al. 2013; Polyak et al. 2015).
Members of the superfamily of heterotrimeric G (guanine–nucleotide binding) protein-coupled cell surface receptors (GPCRs), also called seven transmembrane domain receptors, are activated by various neurotransmitters, including dopamine, serotonin, GABA and glutamate (Geppetti et al. 2015). GPCRs regulate physiological responses through the activation of heterotrimeric G proteins, which are composed of three subunits, α, β and γ. A specific subtype of Gα is i/o [Gα(i/o)], which is implicated in the inhibition of cAMP/PKA (cyclic adenosine monophosphate/protein kinase-A) signalling, thereby depressing certain neuronal activities, such as those involved in reward system, motivation and task salience (Brust et al. 2015).
The nematode Caenorhabditis elegans develops as either a hermaphrodite (with two sex chromosomes, XX) or a male (with only one sex chromosome, XO). The self-fertile hermaphrodite is essentially a female capable of producing sperms for a brief period before oogenesis. In this organism, Gα(i/o) is encoded by goa-1 (G protein, O, alpha subunit) (Mendel et al. 1995), the closest paralogue of which is gpa-16 (G protein, alpha subunit) (Gotta and Ahringer 2001). GOA-1/Gα(i/o) accumulates in all neurons and controls several sex-specific behavioural patterns, including hermaphrodite egg-laying, male mating, locomotion (e.g., males generally move faster than hermaphrodites) and feeding (Mendel et al. 1995; Ségalat et al. 1995). Besides behaviour, GOA-1 controls numerous aspects of neuronal physiology, such as response to volatile anaesthetics, olfactory adaptation and neuronal migration (van Swinderen et al. 2001; Bastiani and Mendel 2006; Yamada et al. 2009).
Somatic sexual fates in C. elegans are specified by the global sex-determination pathway, the terminal effector of which is the transcription factor TRA-1 (sexual transformer) (Fig. 1a) (Zarkower and Hodgkin 1992). TRA-1 is orthologous to Drosophila Ci (Cubitus interruptus) and human GLI (glioma-associated) zinc finger proteins, which serve as downstream transcription factors for the Hedgehog (Hh) signalling pathway. TRA-1 specifies hermaphrodite fates by repressing male-specific genes. Its target genes identified so far include egl-1 (egg-laying defective; it controls sexually dimorphic cell deaths), mab-3 (male abnormal; it governs male development and behaviour), fog-3 (feminization of germline; it determines germ cell fate), ceh-30 (C. elegans homeobox; it protects male-specific neurons from undergoing apoptosis), dmd-3 (doublesex/mab-3 domain family; it regulates sex-specific morphogenesis), xol-1 (XO lethal; the master sex-switch gene), lin-39 (abnormal cell lineage; it controls vulval development), daf-16 (dauer larva formation defective; it is required for longevity and dauer larva development) and unc-6 (uncoordinated; it affects synaptic connectivity) (Conradt and Horvitz 1999; Yi et al. 2000; Chen and Ellis 2000; Schwartz and Horvitz 2007; Mason et al. 2008; Hargitai et al. 2009; Szabó et al. 2009; Berkseth et al. 2013; Hotzi et al. 2018; Weinberg et al. 2018). In this study, we identified a novel TRA-1 target gene, goa-1. We show that TRA-1 binds to a conserved regulatory element in the goa-1 coding region, and lowers neuronal accumulation of GOA-1 in hermaphrodite animals compared to males. TRA-1 affects food leaving, male mating, and adaptation to odorants in a GOA-1-dependent manner.
The goa-1 genomic region contains a conserved, functional TRA-1 binding site. a The C. elegans sex-determination cascade. TRA-1 acts as a terminal transcription factor of the pathway. In hermaphrodites, TRA-1 represses male-specific genes. Bars represent inhibitory interactions, arrows indicate activations. X sex chromosome, A autosome. b The structure of goa-1. Boxes represent exons, connecting lines correspond to introns. The position of the consensus TRA-1 binding site is indicated (red arrowhead). In the binding sequence, red letters indicate strictly conserved nucleotides, while green ones represent conserved nucleotides. The paralogous gpa-16 genomic region significantly differs in sequence from the consensus TRA-1 binding site (non-conserved nucleotides are underlined). Although the paralogous goa-1 and gpa-16 nucleotide sequences differ from each other at this site, the corresponding amino acid sequences they encode are identical. c Multiple sequence alignments of goa-1 (up) and gpa-16 (down) genomic fragments from closely related Caenorhabditis species. Each of the short stretches contains a conserved (or divergent) TRA-1 binding element. This fragment is located within the 5th exon. Red letters: identical nucleotides; green letters: highly conserved nucleotides; black letters: non-conserved nucleotides; grey letters: flanking sequences. Bar indicates the extent of the TRA-1 binding site. The blue background highlights the conserved elements within this regulatory motif. d Electromobility shift assay showing an efficient and specific in vitro binding of TRA-1 to its conserved regulatory element identified within the 5th exon of goa-1 coding region (lane 5). TRA-1 does not bind an oligonucleotide specific to the corresponding paralogous gpa-16 sequence (lane 8). Creating a consensus TRA-1 site in this gpa-16 region establishes binding (lane 11). The binding between TRA-1 and a xol-1-pecific oligo was used as a positive control (lane 3). oligo: oligonucleotide, rest: restored, mut: mutated. Lane 1: no reticulocyte; lanes 2–11: reticulocyte was added. e, f Quantitative real-time PCR analysis demonstrates that transcript levels of goa-1 are fewer in XX hermaphrodites than in XO males and tra-1(-) mutant XX animals. Transcript levels of goa-1 are lower in animals with tra-1(e1575gf/+); tra-3(e1767) genotype than in tra-3(e1767) mutant control animals. Young, non-gravid hermaphrodites were examined. Data were normalized with their own housekeeping gene data, then results were further normalized in a way that each data was divided by the average control [wild-type and tra-3(-) mutant hermaphrodite] data. Bars represent ± SD, *P < 0.05. NS not significant, Mann–Whitney U test P value. Statistics are included in Table S1
Materials and methods
Nematode strains and mutant alleles
Caenorhabditis elegans Bristol (N2) strain was used as wild-type. The following mutant/transgenic strains were used:
-
DG1856 goa-1(sa734)I
-
MT2426 goa-1(n1134)I
-
PS1493 dpy-20(e1362)IV; syIs9[(pMH86)dpy-20(+) + pJMGoQL)]
-
CB2823 tra-1(e1488)III; eDp6(III;f)
-
CB2590 tra-1(e1099)/dpy-18(e1096)III
-
CB3844 fem-3(e2006)IV
-
CB4088 him-5(e1490)V
-
CB3769 tra-1(e1575gf)/+ III; tra-3(e1767)IV
-
TTV393 goa-1(sa734)I; tra-1(e1099)/+ III
-
TTV377 goa-1(n1134)I; tra-1(e1488)III; eDp6(III;f)
-
TTV376 goa-1(n1134)I; him-5(e1490)V
-
TTV366 goa-1(sa734)I; him-5(e1490)V
-
TTV384 dpy-20(e1362)IV; syIs9[(pMH86)dpy-20(+) + pJMGoQL)]; him-5(e1490)V
-
TTV413 unc-119(ed3)III; eluIs307(pgoa-1::mutGOA-1::GFP)
-
TTV414 unc-119(ed-3)III; him-5(e1490)V; eluIs306(pgoa-1::GOA-1::GFP)
-
TTV411 tra-1(e1099)/unc-119(ed-3)III; eluIs306(pgoa-1::GOA-1::GFP)
-
TTV412 fem-3(e2006)IV; unc-119(ed-3); eluIs306 (pgoa-1::GOA-1::GFP)
-
TTV416 unc-119(ed3)III; eluIs306(pgoa-1::GOA-1::GFP)III
-
TTV386 unc-119(ed3)III; eluEx312(goa-1::ppes-10::gfp)
-
TTV396 unc-119(ed3)III; eluEx313(mutgoa-1::ppes-10::gfp)
-
TTV402 unc-119(ed3)III; eluEx319(gpa-16::ppes-10::gfp)
-
TTV407 unc-119(ed3)III; eluEx324(resgpa-16::ppes-10::gfp)
-
TTV655 fem-3(e2006)IV; unc-119(ed-3); eluIs307(pgoa-1::mutGOA-1::GFP)
-
TTV645 tra-1(e1099)/unc-119(ed-3)III; eluIs307(pgoa-1::mutGOA-1::GFP)
-
TTV415 unc-119(ed3)III; him-5(e1490)V; eluIs307(pgoa-1::mutGOA-1::GFP)
-
TTV688 him-5(e1490)V; bxIs14[pkd-2::GFP + pdx-1)]
-
TTV689 goa-1(sa734)I; him-5(e1490)V; bxIs14[pkd-2::GFP + pdx-1(pha-1)]
-
TTV690 goa-1(syIs9)I; him-5(e1490)V; bxIs14[pkd-2::GFP + pdx-1]
-
TTV691 egl-30(ad806)I; him-5(e1490)V
-
BY200 vtIsl(dat-1::GFP; rol-6)
-
TTV746 goa-1(sa734)I; vtIsl
-
TTV747 goa-1(syIs9)I; vtIsl
In vitro DNA–protein binding assay
EMSA (electromobility shift assay) was performed as described (Hargitai et al. 2009). The following forward and reverse oligonucleotides were used: wt (wild-type) xol-1: 5′-GGG GCC CCT GTA AGA CCA CACA CGA AAA CCT CTT GTT-3′ and 5′-GGG GAA CAA GAG GTT TTC GTC GTG TGT GGT CTT ACA GGG G-3′; wt goa-1: 5′-GGG GGT ACA GAT TGT TCG ATG TGG GAG GTC AAA GAT CAG A-3′ and 5′-GGG GGT CTG ATC TTT GAC CTC CCA CAT CGA ACA ATC AGT A-3′; mut (mutated) goa-1: 5′-GGG GGC CCC TGT AAC GGT ACA CAC GAC GAA AAC CTC TTG GGG ACA TT-3′ and 5′-GGG GGA ATG TCC CCA ACA AGA GGT TTT CGT GTG TAC CGT TAC AGG GG-3′; wt gpa-16: 5′-GGG GGT TCA GGG TTT TCG ACG TTG GCG GAC AGC GAT CCG A-3′ and 5′-GGG GGT CGG ATC GCT GTC CGC CAA CGT CGA AAA CCC TGA A-3′; mut gpa-16: 5′-GGG GGT TCA GGG TTT TCG ACG TGG GAG GTC AGC GAT CCG A-3′ and 5′-GGG GGT CGG ATC GCT GAC CTC CCA CGT CGA AAA CCC TGA A-3′.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from 40 synchronized animals per each sample (Fig. 1e, f) or from embryos collected by bleaching gravid hermaphrodites (Fig. S5), using TRI reagent (Sigma, T9424). Total RNA was used for first strand cDNA synthesis by RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, K1622). At embryonic stages, nearly 50% of the “wild-type males” sample, 30% of the “him-5(-) mutants” sample, and only a small fraction of the “tra-1(e1099/+) mutants” sample consist of males. Real-time PCR was performed on LightCycler 96 System (Roche, FastStart Essential DNA Green Master, 06402712001), using the following conditions: denaturation at 95 °C for 10 min, followed by 45 cycles of amplification (10 s, 95 °C; 10 s, 58 °C and 20 s, 72 °C). PCR products were detected by the fluorescence of double strand DNA binding SYBRGreen dye. Melting curve analysis was performed to confirm correct PCR product size and absence of nonspecific bands. Mean relative mRNA levels were determined by normalizing the PCR threshold cycle number of goa-1 with that of cdc-42 and pmp-3 reference genes (Hoogewijs et al. 2008) and using the 2−ΔC(t) method (Pfaffl 2001). Forward and reverse primers were as follows. pmp-3: 5′-TGG AAT TGT TTC ACG GAA TG-3′ and 5′-TCA GCT CTT CGT GAA GTT CC-3′; cdc-42: 5′-TGG AGA GAA GTT GGC AAA GG-3′ and 5′-TGT TGT GGT GGG TCG AGA G-3′; goa-1: 5′-TGG TTC GGC TGA CAG AGA G-3′ and 5′-CCG TTT CAT TGA ACT GAG CA-3′.
Food-leaving (mate searching) assay
Food leaving assays were performed as described (Chasnov et al. 2007). Briefly, plates of 9 cm diameter were filled with 10 ml NGM (nematode growth) agar medium, and dried overnight at RT, then 18 μl Escherichia coli OP50 overnight culture was inoculated in the centre of each plate. Virgin males and hermaphrodites were selected at the L3 larval stage, and grown separately. 20 animals were placed individually on each plate, and scored for leaving for 24 h. Results were compared with two methods. First, Friedman with Wilcoxon Signed Ranks Post Hoc-test was performed on animals remaining on plates. Second, we calculated PL (probability of leaving): N(t)/N(0) = exp(−PLt) (Lipton et al. 2004) for each hour, then averaged data, and finally performed a Kruskal–Wallis H test with Dunn’s Post Hoc-test.
Male mating assay
Male mating assay was performed as described (Lipton et al. 2004). Onto plastic dishes of 3 cm diameter, filled with NGM, 1 cm diameter (~ 16 μl) of E. coli OP50 culture was inoculated. Five young unc-31(-) mutant (paralyzed) hermaphrodites were placed on the bacterial lawn, and one virgin male was placed on the edge of the bacterial lawn and monitored for 10 min. Time was measured when the male touched and found the vulva of the mating partner. Measurements were stopped when the male inserted its spiculum into a hermaphrodite. If males could not find hermaphrodites within 10 min as in the case of goa-1(-) mutants, the former were moved next to the latter by a platinum wire to assay response reaction.
Sex-pheromone chemotaxis assay
Chemotaxis assay was performed as described (Chasnov et al. 2007). 2 μl NaN3 (Sigma) were dropped into two spots each 3 cm apart the centre of the plate and dried. 2–2 μl attractant and control solution were placed on top of the NaN3 spots, then 20 virgin males or adult hermaphrodites were placed on the centre of the slide, and assayed for 30 min. Virgin males and hermaphrodites were selected individually at the L3 larval stage, then grown separately. Sex pheromone was isolated from C. elegans and C. remanei (EM464 strain) by putting 5 young virgin hermaphrodites (C. elegans) and females (C. remanei) into 100 μl M9 solution for 6 h at 25 °C. The supernatant of C. remanei solution was used as attractant and the supernatant of C. elegans was used as control (C. elegans males are attracted by the pheromone of C. remanei females). The assay was performed on microscope slides covered with a layer of 1.5% agar. The chemotaxis index was calculated as [(number of animals within attractant spot)-(number of animals within control spot)]/(total number of animals).
Adaptation to isoamyl alcohol (IAA)
Adaptation assays were modified from Ref. Lipton et al. (2004). Well-fed, age-synchronized animal were washed three times with assay buffer, incubated in adaptation conditions with IAA (pre-exposure) or with EtOH (mock-pre-exposure—control) for 1 h, then washed twice with assay buffer and placed on chemotaxis plates. 1 μl NaN3 (Sigma) was dropped into two spots each one equal distance from the centre of 9-cm assay plate, and 2 μl attractant (IAA—1% final concentration) and 2 μl control (EtOH) were placed on top of the NaN3 spots. Animals were placed on the centre of the plate, and assayed for 1 h. The chemotaxis index was calculated as [(number of animals within attractant spot) − (number of animals within control spot)]/(total number of animals). The rate of adaptation was calculated as [(chemotaxis index for control) − (chemotaxis index for attractant)]/(chemotaxis index for control).
Reporter constructs
The full length goa-1 coding region and 4000 bp upstream regulatory sequences were amplified by Expand Long Template PCR System (Roche), using the following primers: 5′-TTG GCG CGC CCT CGT CCA TAC TCA TTA CAG TTG C-3′ and 5′-ATT TGC GGC CGC CAA TAA TCA CAT CGG TGA CAG C-3′, and cloned with AscI and NotI restriction enzymes (Fermentas) into the vector pRH21 to generate a GFP-fused full length translational reporter construct (pgoa-1::GOA-1::GFP). The construct (eluIs306) was bombarded into unc-119(ed3) mutant young adult hermaphrodites. To delete the putative TRA-1-binding site (eluIs307), Quick Change Site-Directed Mutagenesis kit (Stratagene) was used with the following primers: 5′-GAA TAT TTA CAG ATT GTT CGA TGT AAG ATC AGA AAG GAA GAA GTG GAT-3′ and 5′-ATC CAC TTC TTC CTT TCT GAT CTT ACA TCG AAC AAT CTG TAA ATA TTC-3′ (pgoa-1::mutGOA-1::GFP). To generate a GFP-fused transcriptional fusion goa-1 reporter construct (goa-1::ppes-10::gfp), a genomic region was amplified with the following primers: 5′-CCC AAG CTT GGG TTC CTT GAC GAC CTG GAA AG-3′ and 5′-AAC TGC AGC TCA TCT TCG TGC AAC ACT TG-3′, and cloned into the vector pPD134.96 (provided by Andrew Fire, Stanford University, USA) at the HindIII and KpnI (Fermentas) restriction sites. In this construct called eluEx312, a pes-10 minimal promoter drives transgene expression which is active only during early embryogenesis. To mutate the putative TRA-1-binding site in this reporter, fusion PCR was performed with the following inner primers: 5′-TTG GCG GAC AAA GAT CAG AAA GGA AGA AGT GGA TTC ATT GTT-3′ and 5′-TTC TGA TCT TTG TCC GCC AAC ATC GAA CAA TCT GTA AAT ATT CA-3′. The resulted construct was called eluEx313. For generating a transcriptional fusion, pes-10-driven gpa-16::gfp reporter, a genomic fragment was amplified using the following primers: 5′-CCC AAG CTT GGG GGC CGG CTA CTA TCT GAG C-3′ and 5′-AAC TGC AGA ATC GCA CCG TCT GAC AAT C-3′, and cloned into pPD134.96 (the construct is called eluEx319; gpa-16::ppes-10::gfp). In this construct, 4 nucleotides were changed in order to restore the canonical TRA-1-binding site, using the following inner primers (fusion PCR): 5′-TGG GAG GTC AGC GAT CCG AAC GAA AAA AAG TGG ATT CAT TGC TTC GAA G-3′ and 5′-TTT TTT CGT TCG GAT CGC TGA CCT CCC ACG TCG AAA ACC CTG AAT TTC-3′. The resulted construct was named as eluEx324 (resgpa-16::ppes-10::gfp). Constructs were co-bombarded with pRH21 containing C. briggsae unc-119(+) rescue sequence into unc-119(ed3) mutant young adult hermaphrodites, and transgenic lines were established.
Results
TRA-1 binds a genomic fragment found in the goa-1 coding region
In C. elegans, GOA-1 influences several sex-specific behavioural patterns, such as egg-laying, male mating and locomotion (Mendel et al. 1995; Ségalat et al. 1995). This knowledge prompted us to analyse the sequence of the goa-1 locus to identify potential TRA-1-binding sites (Hargitai et al. 2009). The in silico analysis revealed a highly conserved TRA-1-binding element in the fifth exon of the goa-1 gene, and also uncovered a conserved binding site in the orthologous goa-1 coding region of a closely related nematode species, C. briggsae (Fig. 1b). The presence of conserved control elements for transcription factors in coding sequences appears to be a general feature in both prokaryotic and eukaryotic genomes, and may have contributed to codon choice, thereby influencing protein evolution in different species (Stergachis et al. 2013). The paralogous C. elegans gpa-16 site is slightly divergent from the consensus TRA-1-binding sequence, without affecting the amino acid sequence both proteins share at this position (Fig. 1b). Using an appropriate outgroup, we generated a phylogenetic tree showing the relationship between GOA-1 and GPA-16 in C. elegans and three closely related Caenorhabditis species with sequenced genomes, C. briggsae, C. remanei and C. brenneri (Fig. S1). The tree clearly illustrates that the two proteins represent separated but related subfamilies of Gα(i/o) proteins. To extend the phylogenetic comparisons of goa-1 and gpa-16, we performed a multiple sequence alignment of the two paralogs in these Caenorhabditis species, focusing on short stretches that contain the canonical (goa-1) or divergent (gpa-16) TRA-1-binding element (Fig. 1c). This comparison strengthens the assumption that goa-1, but not gpa-16, is under selection to maintain a TRA-1-binding sequence.
Using an electromobility shift assay (EMSA), we next showed that in vitro synthesized TRA-1 proteins are capable of effectively binding a goa-1-specific oligonucleotide containing the conserved binding motif (lane 5 in Fig. 1d). The binding of TRA-1 to a xol-1-specific oligonucleotide was used as a positive control (lane 3 in Fig. 1d) (Hargitai et al. 2009). When some of the conserved nucleotides in this site were mutated, physical interaction was no longer detected, showing a strict specificity for the protein–DNA binding (lane 7 in Fig. 1d). TRA-1 did also not bind an oligonucleotide specific to the corresponding gpa-16 paralogous region (lane 9 in Fig. 1d). However, TRA-1 interacted with a mutated gpa-16-specific oligonucleotide, in which the paralogous sequence was previously changed to produce a canonical binding element (lane 11 in Fig. 1d). We conclude that TRA-1 is capable of binding a goa-1 exonic sequence in vitro.
Using qRT-PCR, goa-1 transcript levels were determined in hermaphrodites versus males, as well as in TRA-1 deficient (applying e1099 loss-of-function and e1488 reduction-of-function alleles versus wild-type) and hyperactive [tra-1(e1575gain-of-function/+); tra-3(e1767)] mutant animals versus the corresponding controls. According to the results, goa-1 was expressed at significantly higher levels in males than hermaphrodites at adult stages (Fig. 1e and Table S1). goa-1 activity was also significantly elevated in tra-1(e1099) mutant XX animals relative to wild-type hermaphrodites (Fig. 1e and Table S1), but became decreased in the tra-1(e1575gf/+); tra-3(e1767) genetic background compared to control tra-3(e1767) (Fig. 1f and Table S1). These results indicate that TRA-1 represses goa-1 presumably through the conserved binding site identified in the fifth exon. It is worth mentioning that goa-1 was not identified in a previous unbiased ChIP-Seq (chromatin immunoprecipitation assay followed by deep sequencing) study as a TRA-1 target (Berkseth et al. 2013). However, in that analysis, the authors identified 184 potential binding sites for TRA-1, among which several previously described TRA-1 target genes, such as egl-1 (Conradt and Horvitz 1999), ceh-30 (Schwartz and Horvitz 2007) and lin-39 (Szabó et al. 2009), were not represented.
TRA-1 represses the expression of a goa-1 exonic fragment containing a TRA-1-binding element
To assess the in vivo functionality of the TRA-1-binding element we identified in the goa-1 coding region (within the 5th exon), we next generated a gfp (green fluorescent protein)—tagged, heterologous, embryonic stage-specific, minimal promoter (pes-10) -driven goa-1 reporter, goa-1(bs)::ppes-10::gfp (Fig. 2a) (Hope 1991; Seydoux and Fire 1994). The construct contained a short fragment of the fifth exon of goa-1, with the potential TRA-1-binding sequence (bs) (Fig. 1b). goa-1(bs)::ppes-10::gfp expression was effectively repressed in XX embryos in an otherwise wild-type genetic background, but ectopically expressed in XX embryos depleted for TRA-1 (Fig. 2A’–A’’). Thus, TRA-1 is likely to attenuate goa-1 expression through this bs. Furthermore, a mutated version of the reporter, mutgoa-1(-bs)::ppes-10::gfp, which lacks some critical nucleotides in the predicted TRA-1-binding site (grey letters in Fig. 2a), also displayed ectopic expression in XX embryos (Fig. 2A’’’). We conclude that this highly conserved binding element for TRA-1 may mediate the inhibitory effect of TRA-1 on goa-1 transcription.
The expression of goa-1, but not gpa-16, is repressed by TRA-1 throughout embryonic development. a Structure of the heterologous goa-1(bs)::ppes-10::gfp reporter used in this study. Nucleotides that were changed in the mutant version are indicated by underlining. A’, A’’ Expression of goa-1(bs)::ppes-10::gfp in embryos in wild-type (A’; no expression) versus tra-1(RNAi) (A’’; abundant expression) genetic backgrounds. (A’’’) When the TRA-1 binding site was previously mutated [mutgoa-1(-bs)::ppes-10::gfp; right panel], ectopic expression in embryos becomes obvious. Reporter expression, or its absence, was evident at any embryonic stages examined. So, TRA-1 may control goa-1 throughout embryogenesis. b Structure of the gpa-16(-bs)::ppes-10::gfp reporter used in this study. Nucleotides that were changed in the restored binding site version are indicated by underlining. (B’) Embryonic expression of gpa-16(-bs)::ppes-10::gfp in an otherwise wild-type genetic background. (B’’) Generating a consensus TRA-1 binding site [mutgpa-16(bs)::ppes-10::gfp] abolished expression, which was re-established by treatment with tra-1-specific double-stranded RNA (B’’’). In panels A’–A’’’, and B’–B’’’, the small windows show the corresponding Nomarski pictures. Scale bars represent 50 µm. In panels a and b, black boxes indicate exonic sequences, interconnecting lines represent intronic sequences, the blue box displays the pes-1 minimal promoter, the green box correspond to the gfp (green fluorescent protein) coding region, and the red triangle indicates the conserved TRA-1 binding element. bs the presence of the consensus TRA-1 binding site, while “-bs” its absence
We also created a gpa-16(-bs)::ppes-10::gfp reporter and its binding site mutant version, mutgpa-16(bs)::ppes-10::gfp, in which the divergent paralogous site was changed to a canonical bs for TRA-1 to assay the functional independence of TRA-1 on this genomic fragment (Fig. 2b). We found that gpa-16(-bs)::ppes-10::gfp is abundantly expressed in early embryos (shown by white arrows in Figs. 2B’ and S2). Thus, the paralogous gpa-16 exonic sequence is not repressed by TRA-1. However, the generation of a conserved TRA-1-binding sequence at this position in an otherwise gpa-16 genomic environment (underlined red letters in Fig. 2b) completely abolished expression (i.e., the mutated gpa-16 exonic fragment became responsive to TRA-1) (Figs. 2B’’ and S2). Downregulation of tra-1 by RNA interference (RNAi) in embryos transgenic for mutgpa-16(bs)::ppes-10::gfp resettled expression activity (Figs. 2B’’’ and S2). Based on these data, we suggest that TRA-1 influences goa-1 expression through the conserved exonic binding element.
TRA-1 mitigates the accumulation of a translational fusion GOA-1 reporter in hermaphrodite embryos
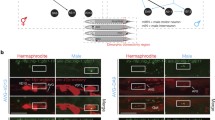
We further tested the in vivo regulatory effect of TRA-1 on goa-1 activity by generating and monitoring the expression of a translational fusion GOA-1::GFP reporter (Fig. 3a) in hermaphrodite vs. male animals, and also in wild-type vs. TRA-1 defective/hyperactive genetic backgrounds. Reporter expression was evident in the nervous system in both sexes at adult stages (Figs. 3b and S3). The fusion protein accumulated at relatively low (basal) levels throughout embryogenesis in wild-type populations, which predominantly consist of hermaphrodite animals (first panel in Fig. 3c and Table S2). In him-5(-) (high incidence of males) mutant embryos, however, nearly 33% of which develop as males as a result of frequent chromosomal non-disjunction during meiosis (Meneely et al. 2012), GOA-1::GFP accumulation appeared at significantly higher levels in a portion of samples that is comparable with the elevated male ratio [Figs. 3c (second panel), C’ and S4A, and Tables S2 and S3]. The quantification of expression intensities revealed that GOA-1 levels are around five times higher in male embryos than in hermaphrodite ones (Fig. 3C’’ and Table S2). We also examined GOA-1::GFP expression in tra-1(-) and fem-3(-) (feminization) mutant embryos, and found that TRA-1 deficiency significantly increases, while TRA-1 hyperactivity (in a FEM-3 defective background) markedly decreases the accumulation of the fusion protein in XX animals, as compared with the wild-type genetic background [Figs. 3C–C’’ (third and fourth panels) and S4A, and Tables S2 and S3]. Thus, goa-1 expression can be decoupled from the number of sex chromosome by modulating TRA-1 activity.
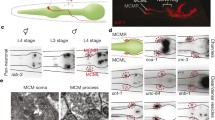
TRA-1 inhibits GOA-1 accumulation during embryonic development. a The structure of the translational fusion GOA-1::GFP reporter used in this study. Black boxes indicate exons, interconnecting lines represent introns, the green box displays gfp (green fluorescent protein) coding region, the red triangle shows the conserved TRA-1 binding element (red, green and blue nucleotides). In the mutated version, the deleted nucleotides are indicated by underlining and grey colouring (the deletion—indicated by a red bar—actually removes 3 amino acids from the protein but does not alter the reading frame). b GOA-1 accumulates in almost all neurons of adult animals (panels show representative GOA-1 accumulation in neurons from the anterior, middle and posterior body parts, respectively). c GOA-1 accumulates at higher levels in male embryos than in hermaphrodite ones (first and second panels). TRA-1 deficiency highly elevates, while TRA-1 hyperactivity (in FEM-3 defective background) decreases GOA-1 accumulation in embryos (third and fourth panels). Embryos with intense GOA-1 accumulation are indicated by red stars. Genotypes are indicated. C’ Percentage of embryos with intense GOA-1 accumulation. Wild-type: embryos mainly consists of XX hermaphrodites (weak expression); him-5(-): nearly 33% of embryos consists of XO males (strong expression); tra-1(e1099/+): only a small fraction of embryos consists of XX animals with male anatomy [theoretically, 25% of progeny derived from a heterozygous tra-1(e1099/+) hermaphrodite should be homozygous mutant and thereby exhibiting male anatomy, but among hermaphrodites chosen randomly for producing embryos only 67% are heterozygous] (only a few embryos with strong expression); fem-3(e2006): embryos are XX female (faint expression). Bars represent ± SEM. C’’ Relative levels of GOA-1 accumulation in a given genotype. Some of the embryos imaged individually were tracked at later stages in life to determine whether they developed as males or hermaphrodites. Their sex and expression intensity corresponded to each other. Bars represent ± SEM, ***P < 0.001; Kruskal–Wallis H test with Dunn’s Post Hoc Test P value. d Expression of a mutGOA-1::GFP reporter, in which the conserved exonic binding site was mutated (see on panel a). Protein accumulation does not respond to mutations altering TRA-1 activity or sex ratio. Expression in each genotype is similar to those found in wild-type embryos. For statistics and the number of animals examined, see Tables S2 and S3
To demonstrate that the modulatory effect of TRA-1 on goa-1 transcription occurs through the conserved binding site we identified in the fifth exon (Fig. 1b), we generated a TRA-1-binding site mutant version of the translational fusion reporter that lack several bases critical for the effective protein–DNA binding (Fig. 3a, d). This mutant reporter (mutGOA-1::GFP) was incapable of responding to mutations that alter him-5 or tra-1 activities (Figs. 3d and S4B, Tables S2 and S3). Rather, expression levels were similar among the wild-type, him-5(-), tra-1(-) and fem-3(-) genetic backgrounds.
We also measured relative goa-1 transcript levels in hermaphrodite vs. male embryos, and wild-type vs. tra-1 defective backgrounds, by using a qRT-PCR analysis. According to the results, goa-1 was expressed at significantly higher levels in males (XO) than in hermaphrodites (XX), and in a tra-1 defective XX background, as compared to the wild type (Fig. S5 and Table S4).
Interestingly, a less significant but still detectable difference in GOA-1 accumulation between hermaphrodites and males could also be observed at larval and adult stages; GOA-1::GFP accumulated at lower levels in hermaphrodite than in male adults (Fig. S6A and Table S5), and similar results were obtained at larval stages (Fig. S7 and Table S6). Consistent with these data, reporter expression was increased in tra-1(-) but decreased in fem-3(-) mutant backgrounds relative to control (Fig. S6B and Table S5). Finally, we also examined the binding site mutant version of the reporter in hermaphrodites versus males (Fig. S6C and Table S5). These data demonstrate that the expression of the mutant reporter does not significantly differ between the two sexes. Together, we conclude that TRA-1 represses goa-1 presumably through the conserved binding element identified in the goa-1 coding region (Fig. 1b), and this regulatory effect is strong during embryogenesis but weak during adulthood.
GOA-1 may influence the specification of male-specific neurons
Compared to the hermaphrodite anatomy, the male nervous system contains 81 additional neurons, and most of the male-specific neurons are located in the tail (Sulston and Horvitz 1977). When the embryo hatches, the hermaphrodite L1 larva contains 222 neurons while the male L1 larva contains 224 neuronal cells. Thus, the majority of male-specific neurons are born during larval development, but some of their precursors (blast cells) may already be specified during embryogenesis.
As goa-1 is expressed at different levels in the two sexes (Figs. 3 and S4, S6), we examined whether goa-1 activity can influence male-specific neuronal development. The protein PKD-2 (human Polycystic Kidney Disease-related) is known to encode a TRPP (transient receptor potential polycystic) cation channel and be expressed in male-specific sensory neurons at both anterior and posterior body regions (four CEMs in the head, and 16 ray B neurons and the HOB in the tail) (Peden and Barr 2005). CEMs are involved in male-specific chemotaxis (sensing hermaphrodites), ray B neurons have a role in mating behaviour, while the HOB participates in the localisation of the hermaphrodite vulva during mating. Using a PKD-2::GFP reporter, we found that pkd-2 is expressed at lower levels in a goa-1 loss-of-function mutant background than in an otherwise wild-type background (Fig. 4 and Table S7). In contrast, goa-1 hyperactivity upregulated pkd-2 expression in male-specific neurons (Fig. 4 and Table S7). Furthermore, a goa-1 gain-of-function mutation could lead to an ectopic expression of pkd-2 in certain (not identified) head neurons of XX hermaphrodites (Fig. S8 and Table S8). Thus, GOA-1 may be involved in the specification (sexual identity) of neurons. To strengthen these results, we examined another marker, dat-1::gfp (vtIs14) (dopamine transporter), which in the tail is expressed only in male-specific neurons (Nass et al. 2002). In the tail of adult males, goa-1 deficiency decreased, while goa-1 hyperactivity slightly increased the expression intensity of dat-1 (Fig. S9 and Table S9). We conclude that GOA-1 may influence neuronal development and functions in a sex-specific manner.
GOA-1 influences pkd-2 expression. pkd-2 is normally expressed in male-specific neurons only. a Accumulation of a PKD-2::GFP fusion protein in head and tail neurons of adult males. PKD-2 accumulates at lower levels in goa-1 defective, but at higher levels in goa-1 hyperactive mutants, as compared to the wild-type background. Images were captured with the same exposure time (40 ms). A’, A’’ Quantification of relative PKD-2 levels in the head (A’) and tail (A’’). Adult animals were examined. Bars represent ± SEM, **P < 0.01, ***P < 0.001; Kruskal–Wallis H test with Dunn’s Post Hoc Test with Bonferroni correction. For statistics, see Table S7
TRA-1 affects behaviour in a GOA-1-dependent manner
If TRA-1 downregulates goa-1 expression during development, particularly throughout embryogenesis, it can be assumed that GOA-1/Gα(i/o)-dependent neuronal functions are influenced by the sex of the animal. To address this issue, we tested behavioural patterns known to be affected by GOA-1/Gα(i/o) in hermaphrodites versus males. In the absence of mating partners, wild-type males leave the food source to find hermaphrodites (Lipton et al. 2004). In contrast, wild-type hermaphrodites do not display such a mate searching behaviour termed food leaving, instead they preferably remain in the bacterial food source. We found that goa-1(-) mutant XO males usually stay within the bacterial lawn, rather than search for hermaphrodites as wild-type males normally do (Fig. 5a and Table S10). Thus, XO males defective for GOA-1 behave as XX hermaphrodites. Similar to wild-type males, goa-1(gf) gain-of-function mutant XX hermaphrodites often left the food source. More significantly, tra-1(-) mutant XX animals that are strongly masculinized are exhibited food-leaving behaviour. Mate searching in tra-1(-) mutants thus was strongly suppressed by GOA-1/Gα(i/o) deficiency (Fig. 5a). This epistatic relationship [the food-leaving phenotype of tra-1(-); goa-1(-) double mutants was identical to that of goa-1(-) single mutants) shows that tra-1 and goa-1 act in the same genetic pathway to control food leaving (tra-1 inhibits goa-1 in the pathway). According to these data, an inactivating mutation in the key sex-determining gene tra-1 decoupled a sex-specific behavioural pattern, mate searching, from the actual karyotype of the animal: tra-1(-) mutants with an XX chromosome set behaved as XO males. Similar to this phenomenon, a loss-of function mutation in goa-1 converted XO males to animals that behave as hermaphrodites, while a hyper-activating goa-1 mutation in XX hermaphrodites rendered a male-like behavioural pattern.
Behaviour and chemosensory information processing in C. elegans are regulated by the TRA-1—goa-1 signalling axis. a Food leaving (mate searching) behaviour in animals with different genetic backgrounds is determined by TRA-1 activity. TRA-1 regulates this trait by inhibiting goa-1. XX hermaphrodites (red circle) remain in, while XO males (dark blue triangle) quickly leave, the food source (bacterial layer) within a couple of hours. GOA-1 deficiency suppresses food leaving behaviour in XO males (light blue cross) and in tra-1(-) mutant XX animals (purple bar). However, goa-1 hyperactivity promotes mate searching (food leaving) in XX hermaphrodites (orange circle). goa-1 gf (hyperactive) mutant XX animals leave the bacterium layer from the onset of adulthood. For statistics, see Table S10. Friedman test (χ2 (9) = 71.79, P < 0.000) with Wilcoxon Signed Ranks Post Hoc Test and Kruskal–Wallis H test with Dunn’s Post Hoc Test. b Chemotactic ability of males to C. remanei female sex pheromone is regulated by TRA-1 and GOA-1. Wild-type XO males (dark blue column) move toward sex pheromone while XX hermaphrodites (purple column) do not. GOA-1 deficiency suppresses chemotaxis to sex pheromone in XO males (light blue column) but not in XX hermaphrodites (yellow column). A hyperactivating mutation in goa-1 elevates the capacity of XX hermaphrodites to move toward sex pheromone (orange column). GOA-1 deficiency (turquoise) suppresses the pheromone attraction phenotype of tra-1(-) mutant XX males (purple). c Adaptation to isoamyl alcohol (IAA) in animals with hyperactive or defective goa-1 function. goa-1(-) mutant hermaphrodites are unable to adapt to IAA whereas goa-1(gf) mutant hermaphrodites showed increased adaptation, as compared with control. fem-3(-) and tra-1(gf) mutant XX animals are also defective for adaptation to IAA, whereas tra-1(-) mutant XX animals with hyperactive GOA-1 function adapt better than control. tra-1(-); goa-1(-) double mutant XX animals are deficient in adaptation. In panel b, results are shown by Box Plot, whereas in panel c, results are shown by Box Plot overlapping Dot Plots. *P < 0.05, **P < 0.01, ***P < 0.001, NS not significant; Kruskal–Wallis H test with Dunn’s Post Hoc-test. For statistics, see Tables S12, S13. d Model showing how behaviour and other neuronal functions in C. elegans are controlled in a sex-specific manner by TRA-1. In this mechanism, TRA-1/GLI, the terminal effector of the sex-determination cascade, directly influences goa-1/Gα(i/o) activity in neurons. GOA-1 influences diverse neuronal functions such as the control of behaviour. Arrows indicate activations, bars represent inhibitory regulatory interactions
We performed a control experiment to find out that changes in mate-searching behaviour (food leaving) in tra-1 and goa-1 mutants are not merely a consequence of altered food perception. To address this issue, egl-30(-) mutant animals were analysed. egl-30, which encodes a Gα(q) protein, antagonizes goa-1 in certain neuronal functions (Matsuki et al. 2006). Mutant males defective for EGL-30 functions quickly left the food source in this assay (Fig. S10 and Table S10). egl-30 loss-of-function mutant XX hermaphrodites at young adult stages also performed a similar food leaving behaviour until they start to lay eggs (Fig. S10 and Table S10) (note that goa-1 gain-of-function mutant XX animals displayed food leaving throughout the entire adulthood; Fig. 5a). We also tested mate-searching behaviour at the L3 larval stage, well before sexual maturation becomes completed. Neither wild-type XO (male) nor goa-1 gf mutant XX (hermaphrodite) animals exhibited a sex drive in food leaving at this larval stage (Fig. S11 and Table S10). Taken together, altering GOA-1 function can decouple a male-specific behavioural pattern, mate searching, from the number of X chromosomes.
When placed in close proximity to hermaphrodites, wild-type males try to rapidly follow and copulate with them (Loer and Kenyon 1993; Liu and Sternberg 1995). This male-specific mating behaviour was effectively blocked by GOA-1/Gα(i/o) deficiency in XO male animals (Fig. S12, and Table S11). tra-1(-) mutant (GOA-1 hyperactive) XX animals developing as males also performed male mating behaviour. GOA-1 deficiency, however, significantly decreased male mating behaviour in XO males and in tra-1(-) mutant XX males (males required more time to copulate). Thus, male mating behaviour in wild-type hermaphrodites is suppressed by the TRA-1-goa-1 regulatory axis. This also indicates that goa-1 functions downstream of tra-1 to suppress male mating behaviour in hermaphrodites.
We assayed the chemotactic ability of hermaphrodites versus males to C. remanei female sex pheromone (Fig. 5b and Table S12). XO males move towards this odorant, while XX hermaphrodites ignore it (Chasnov et al. 2007). GOA-1 deficiency, however, strongly suppressed the capacity of XO males to sense and/or react to the pheromone. Consistent with these data, tra-1(e1099) mutation significantly increased the response of animals with XX karyotype in this chemotaxis assay. goa-1 is hence epistatic to tra-1 in modulating the attraction of tra-1(-) mutant XX animal to sex pheromone. Furthermore, goa-1 hyperactivity likewise elevated the capacity of XX hermaphrodites to move towards sex pheromone (Fig. 5b and Table S12). Taken together, neuronal activity underlying chemotactic behaviour of males to female sex pheromone is affected by both tra-1 and goa-1 activity.
A similar genetic interaction was revealed by monitoring adaptation to another odorant, isoamyl alcohol, in the two sexes (Fig. 5c and Table S13) (Matsuki et al. 2006). Accordingly, mutant nematodes defective for GOA-1 displayed a decreased adaptation to this odorant. In these mutants, reduced tra-1 activity was able to partially rescue adaptation (as more GOA-1 proteins may have been produced). Since e1488 is a reduction-of-function rather than a complete loss-of-function mutation in tra-1, the rescue was only partial. It is possible that in tra-1(e1488) single mutants GOA-1 levels increase by a limited rate only. This led to no significant (detectable) change in adaptation. In goa-1 gain-of-function mutants, however, a much higher GOA-1 activity may exist. Similar to goa-1 inactivating mutations, tra-1 gain-of-function and fem-3 loss-of-function mutations, which lower goa-1 activity, decreased the level of adaptation. Thus, goa-1 plays a role in adaptation to odorants. Because XX hermaphrodites, XO males and tra-1(-) mutant XX animals shared a nearly similar adaptation capacity to isoamyl alcohol, tra-1 is likely to moderately or not affect adaptation in the wild type. In goa-1(-) mutants or in case of tra-1 hyperactivity, tra-1 was capable of modulating the adaptation of animals. We conclude that TRA-1 influences goa-1 in adaptation control even if this genetic interaction occurs at a rather moderate level in the wild-type background. Relative GOA-1 levels, which may depend on TRA-1 activity, determine the physiological outputs of GPCR signalling (Fig. 5d).
GLI proteins, the mammalian orthologues of TRA-1 (Zarkower and Hodgkin 1992, 1993), function as the effectors of an ancient, conserved regulatory circuit, Hh signalling (Briscoe and Thérond 2013). Using a bioinformatical analysis, we found conserved GLI-specific binding sites in the regulatory region of two human Gα(i/o) genes, Gnao1 and Gnai3 (Figs. S13 and S14, and Tables S14 and S15). Although nematodes lack several components of the canonical Hh signalling pathway (Aspöck et al. 1999), and the mechanism of sex-determination in mammals (where sex is determined by the presence of Y chromosome) essentially differs from that found in C. elegans, it is intriguing that Hh signalling also displays numerous gender-specific activities in mammals (Franco and Yao 2012).
Discussion
In this study, we demonstrated that in C. elegans the intracellular accumulation of the heterotrimeric G protein subunit GOA-1/Gα(i/o) is regulated unequally between the two sexes (Figs. 3 and S4, S6). goa-1/Gα(i/o) gene appears to be directly repressed by the major sex-determining factor TRA-1/GLI to specify hermaphrodite-specific behavioural traits and adaptation to odorants (Fig. 5). In silico sequence analysis, in vitro protein–DNA binding assays and in vivo expression data each support the finding that TRA-1 represses goa-1 through a conserved binding site located in the fifth exon of the coding region (Figs. 1, 2, 3). These results imply a direct and specific modulatory effect between the sex-determination machinery and heterotrimeric G protein-mediated signalling in this sexually dimorphic organism. Gα(i/o) is a central component of the GPCR signalling system mediating numerous neuronal functions in divergent animal species (McCudden et al. 2005). If this sex-specific regulatory mechanism involving orthologous TRA-1/GLI proteins and goa-1/Gα(i/o) genes proves to be evolutionally conserved, as suggested by the presence of conserved GLI binding sites in certain GNAI/Gαi loci, especially in GNAO1 (Fig. S14), it could explain some sex-specific differences in human behaviour, chemo-sensation as well as sensitivity to develop Gα(i/o)-related neuropathologies, such as schizophrenia and epilepsy (Bychkov et al. 2011). This could be a realistic mechanism even if primary sex determination is essentially dissimilar between mammals (depends on the presence of chromosome Y) and nematodes (depends on the dosage of chromosome X). One may speculate that the occurrence of chromosome Y somehow lowers Hh signalling in men relative to women, and decreased GLI activity leads to elevated transcript levels of specific GNAI/Gαi genes. As supporting data, Hh signalling in mammals is adjusted unequally in the two sexes, and genes acting in this signalling system participate in sexual differentiation (Franco and Yao 2012).
During embryogenesis, the accumulation of GOA-1 was nearly fivefold greater in males than in hermaphrodites (Fig. 3c, C’’). The protein levels at adult stages, however, were only slightly, but still significantly different in the two sexes (the male:hermaphrodite accumulation ratio was about 3:2; Fig. S6). This characteristic might be explained by other, yet unexplored regulatory factors besides TRA-1, mediating a sex-specific disparity in goa-1 expression. Alternatively, tra-1 activity may display life cycle-specific differences, i.e., TRA-1 may act at higher levels in embryos than in adults. Based on these data, one can speculate that the effect of GOA-1 on behaviour is mainly established during early development.
We also showed that gpa-16, the closest paralogue of goa-1, may be transcribed independently of TRA-1 (Figs. 2 and S2). This may have resulted from sequence divergence in a putative, former TRA-1-binding site in the gpa-16 coding region which presumably occurred after the duplication of the common ancestor of the two genes (Fig. S1). gpa-16 controls cellular processes, such as mitotic spindle positioning and orientation in certain neurons, which display no difference between the sexes (Bastiani and Mendel 2006). goa-1, however, determines behavioural patterns and other neuronal functions unequally in hermaphrodites and males. Hyperactivation of GOA-1, for example, promotes individuals with XX (hermaphrodite) karyotype to behave as males (Fig. 5a, b). Hence, male-specific behavioural traits in this organism are determined by different transcript levels of goa-1, thereby different levels of GPCR-Gα(i/o) signalling, and established through a conserved TRA-1-binding site (Fig. 1). Sex-specific differences in behaviour are a general feature of animal biology. Regulatory interactions between TRA-1/GLI proteins and goa-1/Gα(i/o) genes may be evolutionarily conserved, which can explain the molecular mechanism underlying such a general biological phenomenon.
The expression of several TRA-1 target genes depends entirely on the sex of the animal. For example, xol-1 is expressed only in male but not in hermaphrodite embryos as a primary consequence of the inhibitory effect of signal elements located on chromosome X (SEXs), which is stabilised by TRA-1 repression during later life stages when dosage compensation is already established to reduce the activity sex chromosomes by half (Fig. 1a) (Hargitai et al. 2009). Indeed, xol-1 becomes ectopically active in XX embryos defective for TRA-1. Here, we showed that GOA-1 accumulation highly, but not entirely, relies on TRA-1 activity during embryonic development. As demonstrated by using a translational fusion GFP reporter, GOA-1 abundantly accumulates in male embryos but only at very low levels in hermaphrodite ones (Fig. 3C–C’’). Neurons shared by both sexes are generated at three main developmental periods: first at the proliferation phase of embryogenesis (most neurons are generated at this stage), second at the L1 larval stage, and third at the L2 larval stage (Sulston and Horvitz 1977). Although some of the sex-specific neurons, such as HSNs and CEMs, are also generated at these stages, most of them are born at the late L3/early L4 larval stages (Sulston et al. 1980). It is possible that male-specific neurons that are involved in male mating behaviour and located in the posterior body region may be specified by high levels of goa-1 dosage, which in turn is determined by low TRA-1activity. This way the precisely defined differences in neuroanatomy between the two sexes (hermaphrodites and males have 8 and 91 sex-specific neurons, respectively) may be governed by GOA-1/Gα(i/o). Interestingly, sex differences in GOA-1 accumulation remain even throughout the adulthood, although at a less significant extent as compared with embryonic stages (Fig. S6). Based on these data, we speculate that, as an alternative mechanism, sex-specific neuronal functions in adult nematodes are controlled at least in part by specific dosages of goa-1/Gα(i/o) in a set of neurons.
Until now, TRA-1 was known to determine sexual fates by primarily regulating the activity of intermediate regulatory genes like egl-1 and ceh-30 (cell death), mab-3 and lin-39 (transcription), fog-3 (translation), xol-1 (sex-determination), daf-16 (ageing) and unc-6 (Conradt and Horvitz 1999; Yi et al. 2000; Chen and Ellis 2000; Schwartz and Horvitz 2007; Mason et al. 2008; Hargitai et al. 2009; Szabó et al. 2009; Berkseth et al. 2013; Hotzi et al. 2018; Weinberg et al. 2018). Some of these target genes shape the physical structure of the nervous system itself. Sex-specific differences in C. elegans neuroanatomy can be established by programmed cell death of specific neurons, altered differentiation pattern of neurons or altered synapse pruning (Conradt and Horvitz 1999; Sammut et al. 2015; Oren-Suissa et al. 2016; Fagan et al. 2018). For example, the hermaphrodite-specific neurons (HSNs) that innervate the vulval muscle are generated embryonically in both sexes, but undergo programmed cell death in male larvae. EGL-1, a BH3-containing cell death activator, becomes activated in the male HSNs, but remains repressed by TRA-1 in hermaphrodite larvae. Hence, TRA-1 protects the HSNs in hermaphrodites from undergoing programmed cell death (Conradt and Horvitz 1999). In contrast with the sex-specific maintenance or specification of subsets of neurons or synaptic connections, we showed here that TRA-1 acts directly on a receptor system that is important for specific neuronal functions. goa-1 is active in virtually all neurons of both sexes, and TRA-1 diminishes, but does not completely abolish, GOA-1 accumulation in hermaphrodite animals as compared to males, particularly during embryonic development (Figs. 3 and S2, S4 and S6). We propose that TRA-1 may control the functions of particular classes of neurons through lowering goa-1 transcription. This may represent a novel regulatory mechanism by which the nematode sex-determination machinery establishes neuronal functions unequally between the two sexes.
References
Aspöck G, Kagoshima H, Niklaus G, Bürglin TR (1999) Caenorhabditis elegans has scores of hedgehog-related genes: sequence and expression analysis. Genome Res 9(10):909–923
Bao AM, Swaab DF (2010) Sex differences in the brain, behaviour, and neuropsychiatric disorders. Neuroscientist 16(5):550–565
Bastiani C, Mendel J (2006) Heterotrimeric G proteins in C. elegans. WormBook 13:1–25
Berkseth M, Ikegami K, Arur S, Lieb JD, Zarkower D (2013) TRA-1 ChIP-seq reveals regulators of sexual differentiation and multilevel feedback in nematode sex determination. Proc Natl Acad Sci USA 110(40):16033–16038
Briscoe J, Thérond PP (2013) The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol 14(7):416–429
Brust TF, Conley JM, Watts VJ (2015) Gα(i/o)-coupled receptor-mediated sensitization of adenylyl cyclase: 40 years later. Eur J Pharmacol 763(Pt B):223–232
Bychkov E, Ahmed MR, Gurevich EV (2011) Sex differences in the activity of signalling pathways and expression of G-protein-coupled receptor kinases in the neonatal ventral hippocampal lesion model of schizophrenia. Int J Neuropsychopharmacol 14(1):1–15
Chasnov JR, So WK, Chan CM, Chow KL (2007) The species, sex, and stage specificity of a Caenorhabditis sex pheromone. Proc Natl Acad Sci USA 104(16):6730–6735
Chen P-J, Ellis RE (2000) TRA-1A regulates transcription of fog-3, which controls germ cell fate in C. elegans. Development 127(14):3119–3129
Conradt B, Horvitz RH (1999) The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell 98(3):317–327
Doty RL, Cameron EL (2009) Sex differences and reproductive hormone influences on human odor perception. Physiol Behav 97(2):213–228
Fagan KA, Luo J, Lagiv RC, Schroeder FC, Albrecht DR, Portman DS (2018) A single neuron chemosensory switch determines the valence of sexually dimorphic sensory behaviour. Curr Biol 28(6):902–914
Fattore L, Fratta W (2010) How important are sex differences in cannabinoid action? Br J Pharmacol 160(3):544–548
Franco HL, Yao HH-C (2012) Sex and hedgehog: roles of genes in the hedgehog signalling pathway in mammalian sexual differentiation. Chromosome Res 20(1):247–258
Geppetti P, Veldhuis NA, Lieu T, Bunnett NW (2015) G protein-coupled receptors: dynamic machines for signalling pain and itch. Neuron 88(4):635–649
Gotta M, Ahringer J (2001) Distinct roles for Galpha and Gbetagamma in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat Cell Biol 3(3):297–300
Hargitai B, Kutnyánszky K, Blauwkamp TA, Steták A, Csankovszki G, Takács-Vellai K, Vellai T (2009) xol-1, the master sex-switch gene in C. elegans, is a transcriptional target of the terminal sex-determining factor TRA-1. Development 136(23):3881–3887
Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR (2008) Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol 9:9
Hope IA (1991) ‘Promoter trapping’ in Caenorhabditis elegans. Development 113(2):399–408
Hotzi B, Kosztelnik M, Hargitai B, Takács-Vellai K, Barna J, Bördén K, Málnási-Csizmadia A, Lippai M, Ortutay C, Bacquet C, Pasparaki A, Arányi T, Tavernarakis N, Vellai T (2018) Aging. Cell 17(3):e12724
Lipton J, Kleemann G, Ghosh R, Lints R, Emmons SW (2004) Mate searching in Caenorhabditis elegans: a genetic model for sex drive in a simple invertebrate. J Neurosci 24(34):7427–7434
Liu KS, Sternberg PW (1995) Sensory regulation of male mating behaviour in Caenorhabditis elegans. Neuron 14(1):79–89
Loer CM, Kenyon CJ (1993) Serotonin-deficient mutants and male mating behavior in the nematode Caenorhabditis elegans. J Neurosci 13:5407–5417
Lonsdorf EV, Eberly LE, Pusey AE (2004) Sex differences in learning in chimpanzees. Nature 428(6984):715–716
Mason DA, Rabinowitz JS, Portman DS (2008) dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans. Development 135(14):2373–2382
Matsuki M, Kunitomo H, Iino Y (2006) Goα regulates olfactory adaptation by antagonizing Gqα-DAG signalling in Caenoehabditis elegans. Proc Natl Acad Sci USA 103(4):1112–1117
McCudden CR, Hains MD, Kimple RJ, Siderovski DP, Willard FS (2005) G-protein signalling: back to the future. Cell Mol Life Sci 62(5):551–577
Mendel JE, Korswagen HC, Liu KS, Hajdu-Cronin YM, Simon MI, Plasterk RH, Sternberg PW (1995) Participation of the protein Go in multiple aspects of behaviour in C. elegans. Science 267(5204):1652–1655
Meneely PM, McGovern OL, Heinis FI, Janowitz JL (2012) Crossover distribution and frequency are regulated by him-5 in Caenorhabditis elegans. Genetics 190(4):1251–1266
Nass R, Hall DH, Miller DM, Blakely RD (2002) Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proc Natl Acad Sci USA 99(5):3264–3269
Oren-Suissa M, Bayer EA, Hobert O (2016) Sex-specific pruning of neuronal synapsis in Caenorhabditis elegans. Nature 533(7602):206–211
Peden EM, Barr MM (2005) The KLP-6 kinesin is required for male mating behaviors and polycystin localization in Caenorhabditis elegans. Curr Biol 15(5):394–404
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45
Polyak A, Rosenfeld JA, Girirajan S (2015) An assessment of sex bias in neurodevelopmental disorders. Genome Med 7:94
Sammut M, Cook SJ, Nguyen KC, Felton T, Hall DH, Emmons SW, Poole RJ, Barrios A (2015) Glia-derived neurons required for sex-specific learning in C. elegans. Nature 526(7573):385–390
Schwartz HT, Horvitz RH (2007) The C. elegans protein CEH-30 protects male-specific neurons from apoptosis independently of the Bcl-2 homolog CED-9. Genes Dev 21(23):3181–3194
Ségalat L, Elkes DA, Kaplan JM (1995) Modulation of serotonin-controlled behaviours by Go in Caenorhabditis elegans. Science 267(5204):1648–1651
Seydoux G, Fire A (1994) Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development 120(10):2823–2834
Stergachis AB, Haugen E, Shafer A (2013) Exonic transcription factor binding directs codon choice and affects protein evolution. Science 342(6164):1367–1372
Sulston JE, Horvitz RH (1977) Post-embryonic cell lineages of the nematode Caenorhabditis elegans. Dev Biol 56(1):110–156
Sulston JE, Albertson DG, Thomson JN (1980) The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev Biol 78(2):542–576
Szabó E, Hargitai B, Regos A, Tihanyi B, Barna J, Borsos É, Takács-Vellai K, Vellai T (2009) TRA-1/GLI controls the expression of the Hox gene lin-39 during C. elegans vulval development. Dev Biol 330(2):339–348
van Swinderen B, Metz LB, Shebester LD, Mendel JE, Sternberg PW, Crowder CM (2001) Goalpha regulates volatile anesthetic action in Caenorhabditis elegans. Genetics 158(2):643–655
Weinberg P, Berkshet M, Zarkower D, Hobert O (2018) Sexually dimorphic unc-6/netrin expression controls sex-specific maintenance if synaptic connectivity. Curr Biol 28(4):623–629.e3
Yamada K, Hirotsu T, Matsuki M, Kunitomo H, Iino Y (2009) GPC-1, a G protein gamma-subunit, regulates olfactory adaptation in Caenorhabditis elegans. Genetics 181(4):1347–1357
Yi W, Ross J, Zarkower D (2000) Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behaviour. Development 127(20):4469–4480
Zarkower D, Hodgkin J (1992) Molecular analysis of the C. elegans sex-determining gene TRA-1: a gene encoding two zinc finger proteins. Cell 70(2):237–249
Zarkower D, Hodgkin J (1993) Zinc fingers in sex determination: only one of the two C. elegans TRA-1 proteins binds DNA in vitro. Nucleic Acids Res 21(16):3691–3698
Zuo W, Zhang W, Chen NH (2013) Sexual dimorphism in cerebral ischemia injury. Eur J Pharmacol 711(1–3):73–79
Acknowledgements
Open access funding provided by Eötvös Loránd University (ELTE). This work was supported by the Hungarian Scientific Research Funds OTKA (K109349 and K132439) and MEDinPROT Protein Science Research Synergy Program to TV, and the Advanced Investigator ERC grant “NeuronAge” to NT. AP is supported by a European Commission Marie Curie Actions Programme, Initial Training. Some nematode strains were provided by the CGC (Caenorhabditis Genetics Center), which is founded by NIH Office of Research Infrastructure Programs (P40 OD010440). Funding was provided by VEKOP (Grant no. VEKOP-2.3.2-16-2017-00014).
Author information
Authors and Affiliations
Contributions
VK, BHa, BHo, MK, TK, EG, KB, CO and AP performed experiments; VK, BHa, BHo, MK, CO, AP, NT and TV designed experiments; all authors analysed data; BHa, BHo, NT and TV wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest. The funders provided support in the form of salaries for authors (CO), but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. Although one of the authors (CO) has a commercial affiliation (HiDucator Ltd.), this does not alter our adherence to Molecular Genetics and Geneomics policies on sharing data and materials.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. Procedures involving experimentation on animal subjects are done in accord with the guide of the institution in which the experiments were done.
Additional information
Communicated by Stefan Hohmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kutnyánszky, V., Hargitai, B., Hotzi, B. et al. Sex-specific regulation of neuronal functions in Caenorhabditis elegans: the sex-determining protein TRA-1 represses goa-1/Gα(i/o). Mol Genet Genomics 295, 357–371 (2020). https://doi.org/10.1007/s00438-019-01625-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-019-01625-0