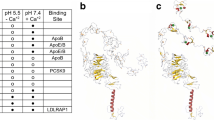
Abstract
Mutations in the low-density lipoprotein receptor (LDLR), which cause familial hypercholesterolemia (FH), present a variable clinical FH phenotype. To date, over 1600 FH-causing mutations have been found worldwide. The aim of this study was to investigate the structure–function relationships of LDLR mutations by using homology modeling. Structural analysis of 36 missense mutations of known receptor activity (33 severe, 1 mild, and 2 non-pathogenic phenotypes) using sequence comparison and homology modeling was performed. Severe phenotypes had less than 2% to 32% of residual LDLR activity. Mild phenotypes had 76–92% of residual LDLR activity. Finally, non-pathogenic phenotypes had normal residual LDLR activity. Sequence comparisons showed that most of the severe phenotypes were located within the fully conserved residues of LDLR, while most of the mild and non-pathogenic phenotypes were located within the poorly conserved residues. Homology modeling demonstrated several phenomena for severe phenotypes: disruption of disulfide bond formation, disturbance of the calcium binding sites, and perturbation of LDLR hydrophobic conserved packing. In contrast, mild and non-pathogenic phenotypes did not disturb the critical region of LDLR. In addition, the root mean square deviation (RMSD) values of severe phenotype tended to be higher than the mild and non-pathogenic phenotypes, and the mean of solvent accessible surface area (ASA) of the residues in wild type structure for the severe phenotype was lower than mild and non-pathogenic phenotypes. These findings provide a better understanding in the structure–function relationships of LDLR mutations and may be useful in predicting FH severity based on future genotyping.







Similar content being viewed by others
Abbreviations
- ASA:
-
Solvent accessible surface area
- CHD:
-
Coronary heart disease
- EGF:
-
Epidermal growth factor
- EGFP:
-
Epidermal growth factor precursor
- FH:
-
Familial hypercholesterolemia
- IMT:
-
Intima-media thickness
- LA:
-
Ligand repeated
- LBD:
-
Ligand-binding domain
- LDL-C:
-
LDL cholesterol
- LDLR:
-
Low density lipoprotein receptor
- OLS:
-
O-linked sugar domain
- PDB:
-
Protein data bank
- RMSD:
-
Root mean square deviation
References
Jeon H, Blacklow SC (2005) Structure and physiology function of the low-density lipoprotein receptor. Annu Rev Biochem 74:535–562
Zou P, Ting AY (2011) Imaging LDL receptor oligomerization during endocytosis using a co-internalization assay. ACS Chem Biol 6(4):308–313
Goldstein JL, Hobbs HH, Brown MS (2001) Familial hypercholesterolemia. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited Disease, 8th edn. McGraw Hill, New York
Francke U, Brown MS, Goldstein JL (1984) Assignment of human gene for the low density lipoprotein receptor to chromosome 19: synteny of a receptor, a ligand and a genetic disease. Proc Natl Acad Sci USA 81:2826–2830
Cuchel M, Bruckert E, Ginsberg HN, Raal FJ, Santos RD, Hegele RA, Kuivenhoven JA, Nordestgaard BG, Descamps OS, Steinhagen-Thiessen E, Tybjaerg-Hansen A, Watts GF, Averna M, Boileau C, Boren J, Catapano AL, Defesche JC, Hovingh GK, Humphries SE, Kovanen PT, Masana L, Pajukanta P, Parhofer KG, Ray KK, Stalenhoef AF, Stroes E, Taskinen MR, Wiegman A, Wiklund O, Chapman MJ (2014) European Atherosclerosis Society Consensus Panel on Familial H: homozygous familial hypercholesterolaemia: new insights and guidance for clinicians to improve detection and clinical management. A position paper from the Consensus Panel on Familial Hypercholesterolaemia of the European Atherosclerosis Society. Eur Heart J 35:2146–2157
British Heart Foundation [Internet database] cited on July 1, 2018. http://www.ucl.ac.uk/ldlr/Current/index.php?select_db=LDLR
Hobbs HH, Brown MS, Goldstein JL (1992) Molecular genetics of the LDL receptor gene in familial hypercholeterolemia. Hum Mutat 1:445–466
Jansen AC, van Wissen S, Defesche JC, Kastelein JJ (2002) Phenotypic variability in familial hypercholesterolaemia: an update. Curr Opin Lipidol 13:165–171
Pimstone SN, Sun XM, du Souich C, Frohlich JJ, Hayden MR, Soutar AK (1998) Phenotypic variation in heterozygous familial Hypercholesterolemia: a comparison of Chinese patients with the same or similar mutations in the LDL receptor gene in China or Canada. Arterioscler Thromb Vasc Biol 18:309–315
Bertolini S, Cassanelli S, Garuti R, Ghisellini M, Simone ML, Rolleri M, Masturzo P, Calandra S (1999) Analysis of LDL receptor gene mutations in Italian patients with homozygous familial hypercholesterolemia. Arterioscler Thromb Vasc Biol 19:408–418
Gudnason V, Day IN, Humphries SE (1994) Effect on plasma lipid levels of different classes of mutations in the low-density lipoprotein receptor gene in patients with familial hypercholesterolemia. Arterioscler Thromb 14:1717–1722
Bertolini S, Cantafora A, Averna M, Cortese C, Motti C, Martini S, Pes G, Postiglione A, Stefanutti C, Blotta I, Pisciotta L, Rolleri M, Langheim S, Ghisellini M, Rabbone I, Calandra S (2000) Clinical expression of familial hypercholesterolemia in clusters of mutations of the LDL Receptor gene that cause a receptor-defective or receptor-negative phenotype. Arterioscler Thromb Vasc Biol 20:E41–E52
Descamps OS, Gilbeau JP, Leysen X, Van Leuven F, Heller FR (2001) Impact of genetic defects on atherosclerosis in patients suspected of familial hypercholesterolaemia. Eur J Clin Invest 31:958–965
Dedoussis GV, Skoumas J, Pitsavos C, Choumerianou DM, Genschel J, Schmidt H, Stefanadis C (2004) FH clinical phenotype in Greek patients with LDL-R defective vs. negative mutations. Eur J Clin Investig 34:402–409
Sun XM, Patel DD, Knight BL, Soutar AK (1998) Influence of genotype at the low density lipoprotein (LDL) receptor gene locus on the clinical phenotype and response to lipid-lowering drug therapy in heterozygous familial hypercholesterolaemia. The Familial Hypercholesterolaemia Regression Study Group. Atherosclerosis 136:175–185
Heath KE, Gudnason V, Humphries SE, Seed M (1999) The type of mutation in the low density lipoprotein receptor gene influences the cholesterol-lowering response of the HMG-CoA reductase inhibitor simvastatin in patients with heterozygous familial hypercholesterolaemia. Atherosclerosis 143:41–54
Brorholt-Petersen JU, Jensen HK, Raungaard B, Gregersen N, Faergeman O (2001) LDL-receptor gene mutations and the hypocholesterolemic response to statin therapy. Clin Genet 59:397–405
Koeijvoets KC, Wiegman A, Rodenburg J, Defesche JC, Kastelein JJ, Sijbrands EJ (2005) Effect of low-density lipoprotein receptor mutation on lipoproteins and cardiovascular disease risk: a parent-offspring study. Atherosclerosis 180:93–99
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23:403–405
Valdar WS (2002) Scoring residue conservation. Proteins 48:227–241
Vriend G (1990) WHATIF: a molecular modeling and drug design program. J Mol Graph 8:52–56
Luethy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with three- dimensional profiles. Nature 356:83–85
Lovell SC, Davis IW, Arendall WB, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC (2003) Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins 50:437–450
Colovos VC, Yeates TO (1993) Verification of protein structures: patterns of non-bonded atomic interactions. Protein Sci 2:1511–1519
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Maiti R, Domselaar GHV, Zhang H, Wishart DS (2004) SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res 32:590–594
Gent J, Braakman I (2004) Low-density lipoprotein receptor structure and folding. Cell Mol Life Sci 61:2461–2470
Jeon H, Meng W, Takagi J, Eck MJ, Springer TA, Blacklow SC (2001) Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair. Nat Struct Biol 8:499–504
Huang JT, Wang MT (2002) Secondary structural wobble: the limits of protein prediction accuracy. Biochem Biophys Res Commun 294:621–625
Imai K, Mitaku S (2005) Mechanisms of secondary structure breakers in soluble proteins. Biophysics 1:55–65
MacArthur MW, Thornton JM (1991) Influence of proline residues on protein conformation. J Mol Biol 218:397–412
Lyu PC, Sherman JC, Chen A, Kallenbach NR (1991) Alpha-helix stabilization by natural and unnatural amino acids with alkyl side chains. Proc Natl Acad Sci USA 88(12):5317–5320
Miyakea Y, Yamamurab T, Sakaic N, Miyataa T, Kokubod Y, Yamamotoa A (2009) Update of Japanese common LDLR gene mutations and their phenotypes: mild type mutation L547V might predominate in the Japanese population. Atherosclerosis 203:153–160
Klee EW, Zimmermann MT (2019) Molecular modeling of LDLR aids interpretation of genomic variants. J Mol Med. https://doi.org/10.1007/s00109-019-01755-3
Guo J, Gao Y, Li X, He Y, Zheng X, Bi J, Hou L, Sa Y, Zhang M, Yin H, Jiang L (2019) Systematic prediction of familial hypercholesterolemia caused by low-density lipoprotein receptor missense mutations. Atherosclerosis 281:1–8
Jeenduang N, Promptmas C, Pongrapeeporn KU, Porntadavity S (2008) Molecular modeling of D151Y and M391T mutations in the LDL receptor. Biochem Biophys Res Commun 377:355–360
Cuesta-Lopez S, Falo F, Sancho J (2007) Computational diagnosis of protein conformational diseases: short molecular dynamics simulations reveal a fast unfolding of r-LDL mutants that cause familial hypercholesterolemia. Proteins 66:87–95
Khan JM, Ranganathan S (2009) A multi-species comparative structural bioinformatics analysis of inherited mutations in α-D-Mannosidase reveals strong genotype-phenotype correlation. BMC Genom 10:S3–S33
Manning JR, Bailey MA, Soares DC, Dunbar DR, Mullins JJ (2010) In silico structure-function analysis of pathological variation in the HSD11B2 gene sequence. Physiol Genom 42(3):319–330
Pey AL, Desviat LR, Gámez A, Ugarte M, Pérez B (2003) Phenylketonuria: genotype-phenotype correlations based on expression analysis of structural and functional mutations in PAH. Hum Mutat 21(4):370–378
Cheng YS, Tang TK, Hwang M (1999) Amino acid conservation and clinical severity of human glucose-6-phosphate dehydrogenase mutations. J Biomed Sci 6(2):106–114
Ferdinandusse S, Ylianttila MS, Gloerich J, Koski MK, Oostheim W, Waterham HR, Hiltunen JK, Wanders RJ, Glumoff T (2006) Mutational spectrum of D-bifunctional protein deficiency and structure-based genotype-phenotype analysis. Am J Hum Genet 78(1):112–124
Tinto N, Zagari A, Capuano M, Simone AD, Capobianco V, Daniele G, Giugliano M, Spadaro R, Franzese A, Lucia Sacchetti L (2008) Glucokinase gene mutations: structural and genotype-phenotype analyses in MODY children from South Italy. PLoS ONE 3(4):e1870. https://doi.org/10.1371/journal.pone.0001870
Saito S, Ohno K, Sugawara K, Sakuraba H (2009) Structural and clinical implications of amino acid substitutions in N-acetylgalactosamine-4-sulfatase: insight into mucopolysaccharidosis type VI. Mol Genet Metab 93(4):419–425
Sukegawa K, Nakamura H, Kato Z, Tomatsu S, Montaño AM, Fukao T, Toietta G, Tortora P, Orii T, Kondo N (2000) Biochemical and structural analysis of missense mutations in N-acetylgalactosamine-6-sulfate sulfatase causing mucopolysaccharidosis IVA phenotypes. Hum Mol Genet 9(9):1283–1290
Rapp C, Bai X (1859) Reithmeier RAF (2017) Molecular analysis of human solute carrier SLC26 anion transporter disease-causing mutations using 3-dimensional homology modeling. Biochim Biophys Acta Biomembr 12:2420–2434
Wacey AI, Krawczak M, Kakkar VV, Cooper DN (1994) Determinations of the factor IX mutational spectrum in haemophilia B: an analysis of missense mutations using a multi-domain molecular model of the activated protein. Hum Genet 94:594–608
Saito S, Ohno K, Okuyama T, Sakuraba H (2016) Structural basis of mucopolysaccharidosis type II and construction of a database of mutant iduronate 2-sulfatases. PLoS ONE 11(10):e0163964
Atkins AR, Brereton IM, Kroon PA, Lee HT, Smith R (1998) Calcium is essential for the structural integrity of the cysteine-rich, ligand-binding repeat of the low-density lipoprotein receptor. Biochemistry 37:1662–1670
Bieri S, Atkins AR, Lee HT, Winzor DJ, Smith R, Kroon PA (1998) Folding, calcium binding, and structural characterization of a concatemer of the first and second ligand-binding modules of the low-density lipoprotein receptor. Biochemistry 37:10994–11002
Guo Y, Yu X, Rihani K, Wang QY, Rong L (2004) The role of a conserved acidic residue in calcium-dependent protein folding for a low density lipoprotein (LDL)-A module implication in structure and function for the LDL receptor superfamily. J Biol Chem 279:16629–16637
Blacklow SC, Kim PS (1996) Protein folding and calcium binding defects arising from familial hypercholesterolemia mutations of the LDL receptor. Nat Struct Biol 3:758–762
North CL, Blacklow SC (1999) Structural independence of ligand-binding modules five and six of the LDL receptor. Biochemistry 38:3926–3935
North CL, Blacklow SC (2000) Evidence that familial hypercholesterolemia mutations of the LDL receptor cause limited local misfolding in an LDL-A module pair. Biochemistry 39:13127–13135
North CL, Blacklow SC (2000) Solution structure of the sixth LDL-A module of the LDL receptor. Biochemistry 39:2564–2571
Varret M, Rabès JP, Collod-Béroud G, Junien C, Boileau C, Béroud C (1997) Software and database for the analysis of mutations in the human LDL receptor gene. Nucleic Acids Res 25(1):172–180
Varret M, Rabés JP, Thiart R, Kotze MJ, Baron H, Cenarro A, Descamps O, Ebhardt M, Hondelijn JC, Kostner GM, Miyake Y, Pocovi M, Schmidt H, Schuster H, Stuhrmann M, Yamamura T, Junien C, Béroud C, Boileau C (1998) LDLR Database (second edition): new additions to the database and the software, and results of the first molecular analysis. Nucleic Acids Res 26(1):248–252
Saha S, Boyd J, Werner JM, Knott V, Handford PA, Campbell ID (2001) Downing AK (2001) Solution structure of the LDL receptor EGF-AB pair: a paradigm for the assembly of tandem calcium binding EGF domains. Structure 9:451–456
Boswell EJ, Jeon H, Blacklow SC, Downing AK (2004) Global defects in the expression and function of the low density lipoprotein receptor (LDLR) associated with two familial hypercholesterolemia mutations resulting in misfolding of the LDLR epidermal growth factor-AB pair. J Biol Chem 279:30611–30621
Ohshiro T, Shimabukuro T, Sunagawa M, Ohta T (2009) An 11-year-old boy with familial hypercholesterolemia showing multiple xanthomas and advanced atherosclerosis, who responded to lipid-lowering therapy using statin. J Atheroscler Thromb 16:698–701
Vieira JR, Whittall RA, Cooper JA, Miller GJ, Humphries SE (2006) The A370T variant (StuI polymorphism) in the LDL receptor gene is not associated with plasma lipid levels or cardiovascular risk in UK men. Ann Hum Genet 70:697–704
Lombardi P, Sijbrands EJ, Kamerling S, Leuven JA, Havekes LM (1997) The T705I mutation of the low density lipoprotein receptor gene (FH Paris-9) does not cause familial hypercholesterolemia. Hum Genet 99:106–107
Heath KE, Whittall RA, Miller GJ, Humphries S (2000) I705 variant in the low density lipoprotein receptor gene has no effect on plasma cholesterol levels. J Med Genet 37:713–715
Graham CA, Wright WT, Mcllhatton BP, Young IS, Nicholls DP (2006) The LDLR variant T705I does not cause the typical phenotype of familial hypercholesterolaemia. Atherosclerosis 188:218–219
Naoumova RP, Neuwirth C, Pottinger B, Whittal R, Humphries SE, Soutar AK (2004) Genetic diagnosis of familial hypercholesterolaemia: a mutation and a rare non-pathogenic amino acid variant in the same family. Atherosclerosis 174:67–71
Chang JH, Pan JP, Tai DY, Huang AC, Li PH, Ho HL, Hsieh HL, Chou SC, Lin WL, Lo E, Chang CY, Tseng J, Su MT, Lee-Chen GJ (2003) Identification and characterization of LDL receptor gene mutations in hyperlipidemic Chinese. J Lipid Res 44:1850–1858
Acknowledgements
This research was partially supported by the New Strategic Research (P2P) project, Walailak University, Thailand.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Porntadavity, S., Jeenduang, N. Structure–Function Relationships of LDL Receptor Missense Mutations Using Homology Modeling. Protein J 38, 447–462 (2019). https://doi.org/10.1007/s10930-019-09860-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-019-09860-5