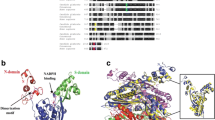
Abstract
3-hydroxy-3-methyl glutaryl-CoA reductase, also known as HMGR, plays a crucial role in regulating cholesterol biosynthesis and represents the main pharmacological target of statins. In mammals, this enzyme localizes to the endoplasmic reticulum membrane. HMGR includes different regions, an integral N-terminal domain connected by a linker-region to a cytosolic C-terminal domain, the latter being responsible for enzymatic activity. The aim of this work was to design a simple strategy for cloning, expression, and purification of the catalytic C-terminal domain of the human HMGR (cf-HMGR), in order to spectrophotometrically test its enzymatic activity. The recombinant cf-HMGR protein was heterologously expressed in Escherichia coli, purified by Ni+-agarose affinity chromatography and reconstituted in its active form. MALDI mass spectrometry was adopted to monitor purification procedure as a technique orthogonal to the classical Western blot analysis. Protein identity was validated by MS and MS/MS analysis, confirming about 82% of the recombinant sequence. The specific activity of the purified and dialyzed cf-HMGR preparation was enriched about 85-fold with respect to the supernatant obtained from cell lysate. The effective, cheap, and easy method here described could be useful for screening statin-like molecules, so simplifying the search for new drugs with hypocholesterolemic effects.





Similar content being viewed by others
References
Goldstein, J. L., & Brown, M. S. (1990). Regulation of the mevalonate pathway. Nature,343, 425–430.
Baigent, C., Keech, A., Kearney, P. M., Blackwell, L., Buck, G., Pollicino, C., et al. (2005). Efficacy and safety of cholesterol-lowering treatment: Prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet,366, 1267–1278.
Brown, M. S., & Goldstein, J. L. (1986). A receptor-mediated pathway for cholesterol homeostasis. Science,232, 34–47.
Stancu, C., & Sima, A. (2001). Statins: Mechanism of action and effects. Journal of Cellular and Molecular Medicine,5, 378–387.
Dolce, V., Cappello, A. R., Lappano, R., & Maggiolini, M. (2011). Glycerophospholipid synthesis as a novel drug target against cancer. Current Molecular Pharmacology,4, 167–175.
Wang, X., Sato, R., Brown, M. S., Hua, X., & Goldstein, J. L. (1994). SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell,77, 53–62.
Tsai, N. W., Lee, L. H., Huang, C. R., Chang, W. N., Chang, Y. T., Su, Y. J., et al. (2014). Statin therapy reduces oxidized low density lipoprotein level, a risk factor for stroke outcome. Critical Care,18, R16.
Li, H., Horke, S., & Forstermann, U. (2013). Oxidative stress in vascular disease and its pharmacological prevention. Trends in Pharmacological Sciences,34, 313–319.
Liao, J. K., & Laufs, U. (2005). Pleiotropic effects of statins. Annual Review of Pharmacology and Toxicology,45, 89–118.
Saeedi Saravi, S. S., Saeedi Saravi, S. S., Arefidoust, A., & Dehpour, A. R. (2017). The beneficial effects of HMG-CoA reductase inhibitors in the processes of neurodegeneration. Metabolic Brain Disease,32, 949–965.
Hamada, M., Sugimoto, M., Matsui, H., Mizuno, T., Shida, Y., Doi, M., et al. (2011). Antithrombotic properties of pravastatin reducing intra-thrombus fibrin deposition under high shear blood flow conditions. Thrombosis and Haemostasis,105, 313–320.
Iannelli, F., Lombardi, R., Milone, M. R., Pucci, B., De Rienzo, S., Budillon, A., et al. (2018). Targeting mevalonate pathway in cancer treatment: Repurposing of statins. Recent Patents on Anti-cancer Drug Discovery,13, 184–200.
Fiorillo, M., Peiris-Pages, M., Sanchez-Alvarez, R., Bartella, L., Di Donna, L., Dolce, V., et al. (2018). Bergamot natural products eradicate cancer stem cells (CSCs) by targeting mevalonate, Rho-GDI-signalling and mitochondrial metabolism. Biochimica et Biophysica Acta,1859, 984–996.
Safwat, S., Ishak, R. A., Hathout, R. M., & Mortada, N. D. (2017). Statins anticancer targeted delivery systems: Re-purposing an old molecule. Journal of Pharmacy and Pharmacology,69, 613–624.
Istvan, E. S., & Deisenhofer, J. (2001). Structural mechanism for statin inhibition of HMG-CoA reductase. Science,292, 1160–1164.
Istvan, E. S., Palnitkar, M., Buchanan, S. K., & Deisenhofer, J. (2000). Crystal structure of the catalytic portion of human HMG-CoA reductase: Insights into regulation of activity and catalysis. The EMBO Journal,19, 819–830.
Luskey, K. L., & Stevens, B. (1985). Human 3-hydroxy-3-methylglutaryl coenzyme A reductase. Conserved domains responsible for catalytic activity and sterol-regulated degradation. The Journal of Biological Chemistry,260, 10271–10277.
du Souich, P., Roederer, G., & Dufour, R. (2017). Myotoxicity of statins: Mechanism of action. Pharmacology & Therapeutics,175, 1–16.
Malachowski, S. J., Quattlebaum, A. M., & Miladinovic, B. (2017). Adverse effects of statins. JAMA,317, 1079–1080.
Alsheikh-Ali, A. A., & Karas, R. H. (2009). The relationship of statins to rhabdomyolysis, malignancy, and hepatic toxicity: Evidence from clinical trials. Current Atherosclerosis Reports,11, 100–104.
Karalis, D. G. (2014). Achieving optimal lipid goals in the metabolic syndrome: A global health problem. Atherosclerosis,237, 191–193.
Joy, T. R., & Hegele, R. A. (2009). Narrative review: Statin-related myopathy. Annals of Internal Medicine,150, 858–868.
Mollace, V., Sacco, I., Janda, E., Malara, C., Ventrice, D., Colica, C., et al. (2011). Hypolipemic and hypoglycaemic activity of bergamot polyphenols: From animal models to human studies. Fitoterapia,82, 309–316.
Gorinstein, S., Leontowicz, H., Leontowicz, M., Krzeminski, R., Gralak, M., Martin-Belloso, O., et al. (2004). Fresh Israeli Jaffa blond (Shamouti) orange and Israeli Jaffa red Star Ruby (Sunrise) grapefruit juices affect plasma lipid metabolism and antioxidant capacity in rats fed added cholesterol. Journal of Agricultural and Food Chemistry,52, 4853–4859.
Cappello, A. R., Dolce, V., Iacopetta, D., Martello, M., Fiorillo, M., Curcio, R., et al. (2016). Bergamot (Citrus bergamia Risso) flavonoids and their potential benefits in human hyperlipidemia and atherosclerosis: An overview. Mini Reviews in Medicinal Chemistry,16, 619–629.
Di Donna, L., Iacopetta, D., Cappello, A. R., Gallucci, G., Martello, E., Fiorillo, M., et al. (2014). Hypocholesterolaemic activity of 3-hydroxy-3-methyl-glutaryl flavanones enriched fraction from bergamot fruit (Citrus bergamia): ‘‘In vivo’’ studies. Journal of Functional Foods,7, 558–568.
Gopal, G. J., & Kumar, A. (2013). Strategies for the production of recombinant protein in Escherichia coli. Protein Journal,32, 419–425.
Snijder, H. J., & Hakulinen, J. (2016). Membrane protein production in E. coli for applications in drug discovery. Advances in Experimental Medicine and Biology,896, 59–77.
Hartley, J. L. (2006). Cloning technologies for protein expression and purification. Current Opinion in Biotechnology,17, 359–366.
Madeo, M., Carrisi, C., Iacopetta, D., Capobianco, L., Cappello, A. R., Bucci, C., et al. (2009). Abundant expression and purification of biologically active mitochondrial citrate carrier in baculovirus-infected insect cells. Journal of Bioenergetics and Biomembranes,41, 289–297.
Salplachta, J., Rehulka, P., & Chmelik, J. (2004). Identification of proteins by combination of size-exclusion chromatography with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and comparison of some desalting procedures for both intact proteins and their tryptic digests. Journal of Mass Spectrometry,39, 1395–1401.
Di Donna, L., Taverna, D., Indelicato, S., Napoli, A., Sindona, G., & Mazzotti, F. (2017). Rapid assay of resveratrol in red wine by paper spray tandem mass spectrometry and isotope dilution. Food Chemistry,229, 354–357.
Persike, M., & Karas, M. (2009). Rapid simultaneous quantitative determination of different small pharmaceutical drugs using a conventional matrix-assisted laser desorption/ionization time-of-flight mass spectrometry system. Rapid Communications in Mass Spectrometry,23, 3555–3562.
Persike, M., Zimmermann, M., Klein, J., & Karas, M. (2010). Quantitative determination of acetylcholine and choline in microdialysis samples by MALDI-TOF MS. Analytical Chemistry,82, 922–929.
Di Donna, L., Benabdelkamel, H., Taverna, D., Indelicato, S., Aiello, D., Napoli, A., et al. (2015). Determination of ketosteroid hormones in meat by liquid chromatography tandem mass spectrometry and derivatization chemistry. Analytical and Bioanalytical Chemistry,407, 5835–5842.
Aiello, D., Cardiano, P., Cigala, R. M., Gans, P., Giacobello, F., Giuffre, O., et al. (2017). Sequestering ability of oligophosphate ligands toward Al3+ in aqueous solution. Journal of Chemical and Engineering Data,62, 3981–3990.
Aiello, D., Furia, E., Siciliano, C., Bongiorno, D., & Napoli, A. (2018). Study of the coordination of ortho-tyrosine and trans-4-hydroxyproline with aluminum(III) and iron(III). Journal of Molecular Liquids,269, 387–397.
Aiello, D., Casadonte, F., Terracciano, R., Damiano, R., Savino, R., Sindona, G., et al. (2016). Targeted proteomic approach in prostatic tissue: A panel of potential biomarkers for cancer detection. Oncoscience,3, 220–241.
Napoli, A., Aiello, D., Aiello, G., Cappello, M. S., Di Donna, L., Mazzotti, F., et al. (2014). Mass spectrometry-based proteomic approach in Oenococcus oeni enological starter. Journal of Proteome Research,13, 2856–2866.
Aiello, D., Materazzi, S., Risoluti, R., Thangavel, H., Di Donna, L., Mazzotti, F., et al. (2015). A major allergen in rainbow trout (Oncorhynchus mykiss): Complete sequences of parvalbumin by MALDI tandem mass spectrometry. Molecular BioSystems,11, 2373–2382.
Aiello, D., Siciliano, C., Mazzotti, F., Di Donna, L., Athanassopoulos, C. M., & Napoli, A. (2018). Molecular species fingerprinting and quantitative analysis of saffron (Crocus sativus L.) for quality control by MALDI 13 mass spectrometry. RSC Advance,8, 36104–36113.
Aiello, D., Giambona, A., Leto, F., Passarello, C., Damiani, G., Maggio, A., et al. (2018). Human coelomic fluid investigation: A MS-based analytical approach to prenatal screening. Scientific Reports,8, 10973.
Li, Y., Cappello, A. R., Muto, L., Martello, E., Madeo, M., Curcio, R., et al. (2018). Functional characterization of the partially purified Sac1p independent adenine nucleotide transport system (ANTS) from yeast endoplasmic reticulum. Journal of Biochemistry,164, 313–322.
Kussmann, M., & Roepstorff, P. (2000). Sample preparation techniques for peptides and proteins analyzed by MALDI-MS. Methods in Molecular Biology,146, 405–424.
Buchner, J., Pastan, I., & Brinkmann, U. (1992). A method for increasing the yield of properly folded recombinant fusion proteins: Single-chain immunotoxins from renaturation of bacterial inclusion bodies. Analytical Biochemistry,205, 263–270.
Bonofiglio, D., Santoro, A., Martello, E., Vizza, D., Rovito, D., Cappello, A. R., et al. (2013). Mechanisms of divergent effects of activated peroxisome proliferator-activated receptor-gamma on mitochondrial citrate carrier expression in 3T3-L1 fibroblasts and mature adipocytes. Biochimica et Biophysica Acta,1831, 1027–1036.
Iacopetta, D., Madeo, M., Tasco, G., Carrisi, C., Curcio, R., Martello, E., et al. (2011). A novel subfamily of mitochondrial dicarboxylate carriers from Drosophila melanogaster: Biochemical and computational studies. Biochimica et Biophysica Acta,1807, 251–261.
Curcio, R., Muto, L., Pierri, C. L., Montalto, A., Lauria, G., Onofrio, A., et al. (2016). New insights about the structural rearrangements required for substrate translocation in the bovine mitochondrial oxoglutarate carrier. Biochimica et Biophysica Acta,1864, 1473–1480.
Lunetti, P., Cappello, A. R., Marsano, R. M., Pierri, C. L., Carrisi, C., Martello, E., et al. (2013). Mitochondrial glutamate carriers from Drosophila melanogaster: Biochemical, evolutionary and modeling studies. Biochimica et Biophysica Acta,1827, 1245–1255.
Vozza, A., De Leonardis, F., Paradies, E., De Grassi, A., Pierri, C. L., Parisi, G., et al. (2017). Biochemical characterization of a new mitochondrial transporter of dephosphocoenzyme A in Drosophila melanogaster. Biochimica et Biophysica Acta,1858, 137–146.
Kurauskas, V., Hessel, A., Ma, P., Lunetti, P., Weinhaupl, K., Imbert, L., et al. (2018). How detergent impacts membrane proteins: Atomic-level views of mitochondrial carriers in dodecylphosphocholine. The Journal of Physical Chemistry Letters,9, 933–938.
Santoro, A., Cappello, A. R., Madeo, M., Martello, E., Iacopetta, D., & Dolce, V. (2011). Interaction of fosfomycin with the glycerol 3-phosphate transporter of Escherichia coli. Biochimica et Biophysica Acta,1810, 1323–1329.
Bolanos-Garcia, V. M., & Davies, O. R. (2006). Structural analysis and classification of native proteins from E. coli commonly co-purified by immobilised metal affinity chromatography. Biochimica et Biophysica Acta,1760, 1304–1313.
Kleinsek, D. A., & Porter, J. W. (1979). An alternate method of purification and properties of rat liver beta-hydroxy-beta-methylglutaryl coenzyme A reductase. The Journal of Biological Chemistry,254, 7591–7599.
Rodwell, V. W., Beach, M. J., Bischoff, K. M., Bochar, D. A., Darnay, B. G., Friesen, J. A., et al. (2000). 3-Hydroxy-3-methylglutaryl-CoA reductase. Methods in Enzymology,324, 259–280.
Zara, V., Dolce, V., Capobianco, L., Ferramosca, A., Papatheodorou, P., Rassow, J., et al. (2007). Biogenesis of eel liver citrate carrier (CIC): Negative charges can substitute for positive charges in the presequence. Journal of Molecular Biology,365, 958–967.
Bonesi, M., Brindisi, M., Armentano, B., Curcio, R., Sicari, V., Loizzo, M. R., et al. (2018). Exploring the anti-proliferative, pro-apoptotic, and antioxidant properties of Santolina corsica Jord. & Fourr. (Asteraceae). Biomedicine & Pharmacotherapy,107, 967–978.
Frattaruolo, L., Carullo, G., Brindisi, M., Mazzotta, S., Bellissimo, L., Rago, V., et al. (2019). Antioxidant and anti-inflammatory activities of flavanones from Glycyrrhiza glabra L. (licorice) leaf phytocomplexes: Identification of licoflavanone as a modulator of NF-kB/MAPK pathway. Antioxidants (Basel),8, 186.
Frimpong, K., Darnay, B. G., & Rodwell, V. W. (1993). Syrian hamster 3-hydroxy-3-methylglutaryl-coenzyme A reductase expressed in Escherichia coli: Production of homogeneous protein. Protein Expression and Purification,4, 337–344.
Polakowski, T., Stahl, U., & Lang, C. (1998). Overexpression of a cytosolic hydroxymethylglutaryl-CoA reductase leads to squalene accumulation in yeast. Applied Microbiology and Biotechnology,49, 66–71.
Ohto, C., Muramatsu, M., Obata, S., Sakuradani, E., & Shimizu, S. (2009). Overexpression of the gene encoding HMG-CoA reductase in Saccharomyces cerevisiae for production of prenyl alcohols. Applied Microbiology and Biotechnology,82, 837–845.
Mayer, R. J., Debouck, C., & Metcalf, B. W. (1988). Purification and properties of the catalytic domain of human 3-hydroxy-3-methylglutaryl-CoA reductase expressed in Escherichia coli. Archives of Biochemistry and Biophysics,267, 110–118.
Song, A. A., Abdullah, J. O., Abdullah, M. P., Shafee, N., Othman, R., Tan, E. F., et al. (2012). Overexpressing 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) in the lactococcal mevalonate pathway for heterologous plant sesquiterpene production. PLoS ONE,7, e52444.
Lukacs, G., Papp, T., Somogyvari, F., Csernetics, A., Nyilasi, I., & Vagvolgyi, C. (2009). Cloning of the Rhizomucor miehei 3-hydroxy-3-methylglutaryl-coenzyme A reductase gene and its heterologous expression in Mucor circinelloides. Antonie van Leeuwenhoek,95, 55–64.
Takahashi, S., Kuzuyama, T., & Seto, H. (1999). Purification, characterization, and cloning of a eubacterial 3-hydroxy-3-methylglutaryl coenzyme A reductase, a key enzyme involved in biosynthesis of terpenoids. Journal of Bacteriology,181, 1256–1263.
Li, J., Xie, Z., Shi, L., Zhao, Z., Hou, J., Chen, X., et al. (2012). Purification, identification and profiling of serum amyloid A proteins from sera of advanced-stage cancer patients. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences,889–890, 3–9.
Easterling, M. L., Colangelo, C. M., Scott, R. A., & Amster, I. J. (1998). Monitoring protein expression in whole bacterial cells with MALDI time-of-flight mass spectrometry. Analytical Chemistry,70, 2704–2709.
Amado, F. M. L., Santana-Marques, M. G., Ferrer-Correia, A. J., & Tomer, K. B. (1997). Analysis of peptide and protein samples containing surfactants by MALDI-MS. Analytical Chemistry,69, 1102–1106.
Cohen, S. L., & Chait, B. T. (1996). Influence of matrix solution conditions on the MALDI-MS. Analysis of peptides and proteins. Analytical Chemistry,68, 31–37.
Zhou, J., & Lee, T. D. (1995). Charge state distribution shifting of protein ions observed in matrix-assisted laser desorption ionization mass spectrometry. Journal of the American Society for Mass Spectrometry,6, 1183–1189.
Frankevich, V., Zhang, J., Dashtiev, M., & Zenobi, R. (2003). Production and fragmentation of multiply charged ions in ‘electron-free’ matrix-assisted laser desorption/ionization. Rapid Communications in Mass Spectrometry,17, 2343–2348.
Lehoux, J. G., Kandalaft, N., Belisle, S., & Bellabarba, D. (1985). Characterization of 3-hydroxy-3-methylglutaryl coenzyme A reductase in human adrenal cortex. Endocrinology,117, 1462–1468.
Bischoff, K. M., & Rodwell, V. W. (1996). 3-Hydroxy-3-methylglutaryl-coenzyme A reductase from Haloferax volcanii: Purification, characterization, and expression in Escherichia coli. Journal of Bacteriology,178, 19–23.
Koh, K. K., Sakuma, I., & Quon, M. J. (2011). Differential metabolic effects of distinct statins. Atherosclerosis,215, 1–8.
Hosomi, N., Kitagawa, K., Nagai, Y., Nakagawa, Y., Aoki, S., Nezu, T., et al. (2018). Desirable low-density lipoprotein cholesterol levels for preventing stroke recurrence: A post hoc analysis of the J-STARS Study (Japan Statin Treatment Against Recurrent Stroke). Stroke,49, 865–871.
Arinze, N., Farber, A., Sachs, T., Patts, G., Kalish, J., Kuhnen, A., et al. (2018). The effect of statin use and intensity on stroke and myocardial infarction after carotid endarterectomy. Journal of Vascular Surgery,68, 1398–1405.
Bolego, C., Poli, A., Cignarella, A., Catapano, A. L., & Paoletti, R. (2002). Novel statins: Pharmacological and clinical results. Cardiovascular Drugs and Therapy,16, 251–257.
McTaggart, F., Buckett, L., Davidson, R., Holdgate, G., McCormick, A., Schneck, D., et al. (2001). Preclinical and clinical pharmacology of Rosuvastatin, a new 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitor. The American Journal of Cardiology,87, 28B–32B.
Acknowledgements
This work was supported by Associazione Italiana per la Ricerca sul Cancro (FG Grant No. 15404/2014) and by Ministero Italiano della Ricerca e dell’Università (MIUR–PRIN 2015, Prot. 201545245K_002).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Curcio, R., Aiello, D., Vozza, A. et al. Cloning, Purification, and Characterization of the Catalytic C-Terminal Domain of the Human 3-Hydroxy-3-methyl glutaryl-CoA Reductase: An Effective, Fast, and Easy Method for Testing Hypocholesterolemic Compounds. Mol Biotechnol 62, 119–131 (2020). https://doi.org/10.1007/s12033-019-00230-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-019-00230-1