Abstract
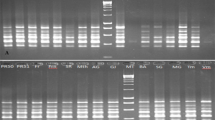
Plums (Prunus spp.) are among the first fruit tree species that attracted human interest. Artificial crosses between wild and domesticated species of plums are still paving the way for creation of new phenotypic variability. In Tunisia, despite a considerable varietal richness of plum as well as a high economic value, the plum sector is experiencing a significant regression. The main reason of this regression is the absence of a national program of plum conservation. Hence, this work was aimed to phenotypically and genetically characterize 23 Tunisian plum accessions to preserve this patrimony. Closely related Prunus species from the same subgenus may be differing at two characteristics: ploidy level and phenotypic traits. In this study, single sequence repeat (SSR) markers allowed distinguishing between eighteen diploid accessions and five polyploid accessions, but SSR data alone precluded unambiguous ploidy estimation due to homozygosity. In contrast, S-allele markers were useful to identify the ploidy level between polyploid species, but they did not distinguish species with the same ploidy level. Seven out of 12 phenotypic traits were shown to be discriminant traits for plum species identification. Molecular and phenotypic traits were significantly correlated and revealed a powerful tool to draw taxonomic and genotypic keys. The results obtained in this work are of great importance for local Tunisian plum germplasm management.







Similar content being viewed by others
Abbreviations
- DNA:
-
Desoxyribonucleic acid
- GSI:
-
Gametophytic self-incompatibility system
- PCR:
-
Polymerase chain reaction
- SFB:
-
S-Haplotype-specific F-box
- S-RNase:
-
S-Ribonuclease
- UPOV:
-
Union internationale de la Protection des Obtentions Variétales
References
Abdallah D, Baraket G, Ben Tamarzizt H, Ben Mustapha S, Salhi-Hannachi A (2016) Identification, evolutionary patterns and intragenic recombination of the gametophytic self incompatibility pollen gene (SFB) in Tunisian Prunus species (Rosaceae). Plant Mol Biol Rep 34:339–352
Arús P, Messeguer R, Viruel F, Tobutt K, Dirlewanger E, Santi F, Quarta R, Ritter E (1994) The European Prunus mapping Project 1994b. In: Schmidt H, Kellerhals M (eds) Progress in temperate furit breeding. Kluwer Academic Publishers, Amsterdam, pp 305–308
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32(3):314–331
Browicz K, Zohary D (1996) The genus Amygdalus L. (Rosaceae): species relationships, distribution and evolution under domestication. Genet Resour Crop Evol 43:229–247
Carrière EA (1872) Amygdalopsis lindleyi. Rev Hortic 1872:33–34
Cipriani G, Lot G, Huang WG, Marrazzo MT, Peterlunger E, Testolin R (1999) AC/GT and AG/CT microsatellite repeats in peach [Prunus persica (L) Batsch]: isolation, characterisation and cross-species amplification in Prunus. Theor Appl Genet 99:65–72
Darlington CD, Ammal Janaki EK (1945) Chromosome atlas of flowering plants. Allen Unwin LTD, London
Das B, Ahmed N, Singh P (2011) Prunus diversity—early and present development: a review. Int J Biodivers Conserv 14:721–734
Dirlewanger E, Cosson P, Tavaud M, Aranzana MJ, Poizat C, Zanetto A, Arùs P, Laigret F (2002) Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Theor Appl Genet 105:127–138
Doyle JJ, Doyle JL (1987) Isolation of DNA from fresh plant tissue. Focus 12:13–15
El Dabbagh N (2016) Analyse de la diversité de processus de développement racinaire chez les Prunus. Aptitude au bouturage et réponses à la contrainte hydrique. Science agricoles. Université d‘Avignon, France
Eldridge L, Ballard R, Baird WV, Abbot A, Morgens P, Callahan A, Scorza R, Monet R (1992) Application of RFLP analysis to genetic linkage mapping in peaches. Sci Hortic 27:160–163
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50
FAOstat (2016) Food and agriculture organization of the United Nations. 798 FAO Statistics Division
Fernández-Cruz J, Fernández-López J, Miranda-Fontaíña ME, Díaz Vazquez R, Toval G (2014) Molecular characterization of Spanish Prunus avium plus trees. For Syst 23:120–128
Galli ZS, Halàsz G, Kiss E, Heszky L, Dobrànszki J (2005) Molecular identification of commercial apple cultivars with microsatellite markers. Sci Hortic 40:1974–1977
García-Verdugo D, Calleja JA, Vargas P, Silva L, Moreira O, Pulido F (2013) Polyploidy and microsatellite variation in the relict tree Prunus lusitanica L.: how effective are refugia in preserving genotypic diversity of clonal taxa? Mol Ecol 22:1546–1557
Gu CZ, Bartholomew B (2003) Prunus Linnaeus. Flora China 9:401–403
Halász J, Makovics-Zsoha N, Szöke F, Ercilis S, Hegedüs A (2017) Simple sequence repeat and S-locus genotyping to explore genetic variability in polyploid Prunus spinosa and P. insititia. Biochem Genet 55:22–33
Hammer O, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:9
Janick J (2005) The origins of fruits, fruit growing, and fruit breeding. Plant breeding reviews. Wiley, New York, pp 255–321
Janick J, Moore JN (1975) Advances in fruit breeding. Purdue University Press, West Lafayette
Kao TH, Tsukamoto T (2004) The molecular and genetic bases of SRNase-based self-incompatibility. Plant Cell 16:72–83
Khadivi-Khub A, Anjam K (2014) Morphological characterization of Prunus scoparia using multivariate analysis. Plant Syst Evol 300:1361–1372
Kota-Dombrovska I, Lācis G (2013) Evaluation of selfincompatibility locus diversity of domestic plum (Prunus domestica L.) using DNA-based S-genotyping. Proc Latv Acad Sci Sect B 67(2):109–115
Lee S, Wen J (2001) A phylogenetic analysis of Prunus and the Amygdaloideae (Rosaceae) using ITS sequences of ribosomal DNA. Am J Bot 88:150–160
Li Y, Smith T, Liu CJ, Awasthi N, Yang J, Wang YF, Li CS (2011) Endocarps of Prunus (Rosaceae: Prunoideae) from the early Eocene of Wutu, Shandong Province, China. Taxon 60:555–564
Linnaeus C (1753) Species plantarum. Salvius, Stockholm
Liu WS, Liu DC, Feng CJ, Zhang AM, Li SH (2005) Genetic diversity and phylogenetic relationships in plum germplasm resources revealed by RAPD markers. J Hortic Sci Biotechnol 81:242–250
Mestre L, Reig G, Betrán J, Moreno MA (2017) Influence of plum rootstocks on agronomic performance, leaf mineral nutrition and fruit quality of ‘Catherina’ peach cultivar in heavy-calcareous soil conditions. Span J Agric Res 15:e0901
Mnejja M, Garcia J, Howad W, Badenes ML, Arùs P (2004) Simple-sequence repeat (SSR) markers of Japanese plum (Prunus salicina Lindl.) are highly polymorphic and transferable to peach and almond. Mol Ecol Notes 4:163–166
Mnejja M, Garcias J, Howad W, Arùs P (2005) Development and transportability across Prunus species of 42 polymorphic almond microsatellites. Mol Eco Notes 5:531–535
Mowrey BD, Werner DJ (1990) Phylogenetic relationships among species of Prunus as inferred by isoenzyme markers. Theor Appl Genet 80:129–133
Nabli M (2011) La flore de la Tunisie, Mise à jour 2011
OCDE (2002) Consensus document on the biology of Prunus spp. (stone fruits), Environment Directorate. Org. Eco. Co-op. and Dev. Paris, pp 1–42
Rehder A (1940) Manual of cultivated trees and shrubs hardy in North America. Exclusive of the subtropical and warmer temperate regions, 2nd edn. MacMillan, New York
Reig G, Jiménez S, Mestre L, Font iForcada C, Betrán JA, Moreno MA (2018) Horticultural, leaf mineral and fruit quality traits of two ‘Greengage’ plum cultivars budded on plum based rootstocks in Mediterranean conditions. Sci Hortic 232:84–91
Salazar JA, Ruiz D, Campoy JA, Sanchez-Perez R, Crisosto CH, Martinez-Garcia PJ, Blenda A, Jung S, Main D, Martinez-Gomez P, Rubio M (2014) Quantitative trait loci (QTL) and mandelian trait loci (MTL) analysis in Prunus: a breeding perspective and beyond. Plant Mol Biol Rep 32:1–18
Sonneveld T, Tobutt KR, Vaughan SP, Robbins TP (2005) Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S-haplotype-specific F-box gene. Plant Cell 17:37–51
Sosinski B, Gannavarapu M, Hager LD, Beck LE, King GJ, Ryder CD, Rajapakse S, Baird WV, Ballard RE, Abbott AG (2000) Characterization of microsatellite markers in peach [Prunus persica (L.) Batsch]. Theor Appl Genet 101:421–428
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tao R, Yamane H, Sugiura A, Murayama H, Sassa H, Mori H (1999) Molecular typing of S-alleles through identification, characterization and cDNA cloning for S-RNases in sweet cherry. J Am Soc Hortic Sci 124:224–233
UPOV (2014) Union Internationale pour la Protection des Obtentions Végétales. Code UPOV: PRUNU_SAL. Genève. www.upov.int
Vaughan SP, Russell K, Sargent DJ, Tobutt KR (2006) Isolation of S-locus F-box alleles in Prunus avium and their application in a novel method to determine self-incompatibility genotype. Theor Appl Genet 112:856–866
Wünsch A, Carrera M, Hormaza JI (2006) Molecular characterization of local Spanish peach [Prunus persica (L.) Batsch] germplasm. Genet Resour Crop Evol 53:925–932
Yamane H, Tao R, Sugiura A, Hauck HR, Iezzoni AF (2001) Identification and characterization of S-RNases in tetraploid sour cherry (Prunus cerasus). J Am Soc Hortic Sci 126:661–667
Zeinalabedini M, Majourhat K, Khayam-Nekoui M, Grigorian V, Torchi M, Dicenta F, Martinez-Gomez P (2008) Comparison of the use of morphological, protein and DNA markers in the genetic characterization of Iranian wild Prunus species. Sci Hortic 16:80–88
Zhang SY (1992) Systematic wood anatomy of the Rosaceae. Blumea 37:81–158
Zhengy W, Raven P, Deyuan H (2003) Flora of China, vol 9. Science Press, Beijing
Zohary D, Hopf M, Weiss E (2013) Domestication of plants in the Old World: the origin and spread of domesticated plants in Southwest Asia, Europe, and the Mediterranean Basin, 4th edn. OUP, Oxford
Acknowledgements
This research was financed by the Tunisian ‘Ministère de l’Enseignement supérieur et de la Recherche Scientifique’. We would like to gratefully thank Tunisian farmers for kindly providing plant material and for their help during phenotypic measurements in the fields. We wish to thank the anonymous reviewers and the editors for their critical comments which allowed us to improve the original manuscript.
Author information
Authors and Affiliations
Contributions
GB performed the experiments and statistical analyses, developed the genetic analyses and wrote the manuscript. DA provided some plant material discussed and corrected the content. SBM provided some plant material and discussed the content. HBT gave additional information regarding plum and corrected the content. ASH offered experimental instructions, supervised and provided editorial advice.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Baraket, G., Abdallah, D., Ben Mustapha, S. et al. Combination of Simple Sequence Repeat, S-Locus Polymorphism and Phenotypic Data for Identification of Tunisian Plum Species (Prunus spp.). Biochem Genet 57, 673–694 (2019). https://doi.org/10.1007/s10528-019-09922-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-019-09922-4