Abstract
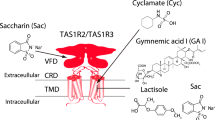
The structure of sweet taste receptor (STR), a heterodimer of class C G-protein coupled receptors comprising T1R2 and T1R3 molecules, is still undetermined. In this study, a new enhanced model of the receptor is introduced based on the most recent templates. The improvement, stability, and reliability of the model are discussed in details. Each domain of the protein, i.e., VFTM, CR, and TMD, were separately constructed by hybrid-model construction methods and then assembled to build whole monomers. Overall, 680 ns molecular dynamics simulation was performed for the individual domains, the whole monomers and the heterodimer form of the VFTM orthosteric binding site. The latter’s structure obtained from 200 ns simulation was docked with aspartame; among various binding sites suggested by FTMAP server, the experimentally suggested binding domain in T1R2 was retrieved. Local three-dimensional structures and helices spans were evaluated and showed acceptable accordance with the template structures and secondary structure predictions. Individual domains and whole monomer structures were found stable and reliable to be used. In conclusion, several validations have shown reliability of the new and enhanced models for further molecular modeling studies on structure and function of STR and C GPCRs.





Similar content being viewed by others
Data Availability and Materials
The datasets generated or analyzed during the current study are not publicly available due to data privacy reasons but are available from the corresponding author on reasonable request.
Abbreviations
- STR:
-
Human sweet taste receptor
- C GPCR:
-
Class C of G-protein coupled receptors
- 7TM:
-
Seven transmembrane α-helices
- VFTM:
-
Venus flytrap domain or module
- CR:
-
Cysteine-rich domain
- TMD:
-
Transmembrane domain
- mGluR1:
-
Metabotropic glutamate receptor subtype 1
- MD:
-
Molecular dynamics
- N-model:
-
The new model introduced in the current study
- R-model:
-
The most recent model presented
- I-model:
-
Model represented by I-TASSER or GPCR-I-TASSER
- SS:
-
Secondary structure
- ns:
-
Nanoseconds
- PME:
-
Particle mesh Ewald
- RMSD:
-
Root mean square deviation
- CA:
-
Carbon-alpha
- SD:
-
Standard deviation
- SDR:
-
Standard deviation ratio
References
Nelson, G., Hoon, M. A., Chandrashekar, J., Zhang, Y., Ryba, N. J., & Zuker, C. S. (2001). Mammalian sweet taste receptors. Cell, 106(3), 381–390.
Li, X., Staszewski, L., Xu, H., Durick, K., Zoller, M., & Adler, E. (2002). Human receptors for sweet and umami taste. Proceedings of the National Academy of Sciences, 99(7), 4692–4696. https://doi.org/10.1073/pnas.072090199.
Behrens, M., & Meyerhof, W. (2011). Gustatory and extragustatory functions of mammalian taste receptors. Physiology and Behavior, 105(1), 4–13. https://doi.org/10.1016/j.physbeh.2011.02.010.
Laffitte, A., Neiers, F., & Briand, L. (2014). Functional roles of the sweet taste receptor in oral and extraoral tissues. Current Opinion in Clinical Nutrition and Metabolic Care, 17(4), 379–385. https://doi.org/10.1097/mco.0000000000000058.
Venkatakrishnan, A. J., Deupi, X., Lebon, G., Tate, C. G., Schertler, G. F., & Babu, M. M. (2013). Molecular signatures of G-protein-coupled receptors. Nature, 494(7436), 185–194.
Pierce, K. L., Premont, R. T., & Lefkowitz, R. J. (2002). Seven-transmembrane receptors. Nature Reviews Molecular Cell Biology, 3(9), 639–650.
Muto, T., Tsuchiya, D., Morikawa, K., & Jingami, H. (2007). Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proceedings of the National Academy of Sciences, 104(10), 3759–3764.
Kniazeff, J., Prézeau, L., Rondard, P., Pin, J.-P., & Goudet, C. (2011). Dimers and beyond: the functional puzzles of class C GPCRs. Pharmacology and Therapeutics, 130(1), 9–25. https://doi.org/10.1016/j.pharmthera.2011.01.006.
Congreve, M., Doré, A.S., Jazayeri, A., Nonoo, R. (2015). Engineering G protein-coupled receptors for drug design. In G. Scapin, D. Patel, E. Arnold (Eds), Multifaceted roles of crystallography in modern drug discovery. (pp 1–18). Netherlands: Springer. https://doi.org/10.1007/978-94-017-9719-1.
Hillisch, A. (2004). Utility of homology models in the drug discovery process. Drug Discovery Today., 9, 659–69.
Cui, M., Jiang, P., Maillet, E., Max, M., Margolskee, R. F., & Osman, R. (2006). The heterodimeric sweet taste receptor has multiple potential ligand binding sites. Current Pharmaceutical Design, 12(35), 4591–4600. https://doi.org/10.2174/138161206779010350.
Xu, H., Staszewski, L., Tang, H., Adler, E., Zoller, M., & Li, X. (2004). Different functional roles of T1R subunits in the heteromeric taste receptors. Proceedings of the National Academy of Sciences of the United States of America, 101(39), 14258–14263. https://doi.org/10.1073/pnas.0404384101.
Servant, G., Tachdjian, C., Tang, X.-Q., Werner, S., Zhang, F., Li, X., Kamdar, P., Petrovic, G., Ditschun, T., Java, A., Brust, P., Brune, N., DuBois, G. E., Zoller, M., & Karanewsky, D. S. (2010). Positive allosteric modulators of the human sweet taste receptor enhance sweet taste. Proceedings of the National Academy of Sciences of the United States of America, 107(10), 4746–4751. https://doi.org/10.1073/pnas.0911670107.
Kim, S.-K., Chen, Y., Abrol, R., Goddard, W. A., & Guthrie, B. (2017). Activation mechanism of the G protein-coupled sweet receptor heterodimer with sweeteners and allosteric agonists. Proceedings of the National Academy of Sciences, 114(10), 2568–2573. https://doi.org/10.1073/pnas.1700001114.
Jiang, P. H., Cui, M., Zhao, B. H., Snyder, L. A., Benard, L. M. J., Osman, R., Max, M., & Margolskee, R. F. (2005). Identification of the cyclamate interaction site within the transmembrane domain of the human sweet taste receptor subunit T1R3. Journal of Biological Chemistry, 280(40), 34296–34305. https://doi.org/10.1074/jbc.M505255200.
Jiang, P., Cui, M., Zhao, B., Liu, Z., Snyder, L. A., Benard, L. M., Osman, R., Margolskee, R. F., & Max, M. (2005). Lactisole interacts with the transmembrane domains of human T1R3 to inhibit sweet taste. The Journal of Biological Chemistry, 280(15), 15238–15246. https://doi.org/10.1074/jbc.M414287200.
Winnig, M., Bufe, B., Kratochwil, N. A., Slack, J. P., & Meyerhof, W. (2007). The binding site for neohesperidin dihydrochalcone at the human sweet taste receptor. BMC Structural Biology, 7, 66 https://doi.org/10.1186/1472-6807-7-66.
Zhang, F., Klebansky, B., Fine, R. M., Liu, H., Xu, H., Servant, G., Zoller, M., Tachdjian, C., & Li, X. (2010). Molecular mechanism of the sweet taste enhancers. Proceedings of the National Academy of Sciences of the United States of America, 107(10), 4752–4757. https://doi.org/10.1073/pnas.0911660107.
Assadi-Porter, F. M., Maillet, E. L., Radek, J. T., Quijada, J., Markley, J. L., & Max, M. (2010). Key amino acid residues involved in multi-point binding interactions between brazzein, a sweet protein, and the T1R2-T1R3 human sweet receptor. Journal of Molecular Biology, 398(4), 584–599. https://doi.org/10.1016/j.jmb.2010.03.017.
Masuda, K., Koizumi, A., Nakajima K-i, Tanaka, T., Abe, K., Misaka, T., & Ishiguro, M. (2012). Characterization of the modes of binding between human sweet taste receptor and low-molecular-weight sweet compounds. PLoS ONE, 7(4), e35380 https://doi.org/10.1371/journal.pone.0035380.
Maillet, E. L., Cui, M., Jiang, P., Mezei, M., Hecht, E., Quijada, J., Margolskee, R. F., Osman, R., & Max, M. (2015). Characterization of the binding site of aspartame in the human sweet taste receptor. Chemical Senses, 40(8), 577–586. https://doi.org/10.1093/chemse/bjv045.
Mayank, JaitakV. (2015). Interaction model of steviol glycosides from Stevia rebaudiana (Bertoni) with sweet taste receptors: a computational approach. Phytochemistry, 116(1), 12–20. https://doi.org/10.1016/j.phytochem.2015.05.006.
Shrivastav, A., & Srivastava, S. (2013). Human sweet taste receptor: complete structure prediction and evaluation. International Journal of Chemical and Analytical Science, 4(1), 24–32.
Yousif, R.H., & Khairudin, N.B.A. (2014). Homology modeling of human sweet taste receptors: T1R2-T1R3. Journal of Medical and Bioengineering, 3(2), 84–86.
Chéron, J.-B., Golebiowski, J., Antonczak, S., & Fiorucci, S. (2017). The anatomy of mammalian sweet taste receptors. Proteins: Structure, Function, and Bioinformatics, 85(2), 332–341. https://doi.org/10.1002/prot.25228.
Nuemket, N., Yasui, N., Kusakabe, Y., Nomura, Y., Atsumi, N., Akiyama, S., Nango, E., Kato, Y., Kaneko, M. K., Takagi, J., Hosotani, M., & Yamashita, A. (2017). Structural basis for perception of diverse chemical substances by T1r taste receptors. Nature Communications, 8, 15530 https://doi.org/10.1038/ncomms15530.
Geng, Y., Mosyak, L., Kurinov, I., Zuo, H., Sturchler, E., Cheng, T. C., Subramanyam, P., Brown, A. P., Brennan, S. C., Mun H-c, Bush, M., Chen, Y., Nguyen, T. X., Cao, B., Chang, D. D., Quick, M., Conigrave, A. D., Colecraft, H. M., McDonald, P., & Fan, Q. R. (2016). Structural mechanism of ligand activation in human calcium-sensing receptor. eLife, 5, e13662 https://doi.org/10.7554/eLife.13662.
Consortium, T. U. (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Research, 45(D1), D158–D169. https://doi.org/10.1093/nar/gkw1099.
Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25. https://doi.org/10.1093/nar/25.17.3389
Krieger, E., & Vriend, G. (2014). YASARA View—molecular graphics for all devices—from smartphones to workstations. Bioinformatics, 30(20), 2981–2982.
Laskowski, R. A., MacArthur, M. W., Moss, D. S., & Thornton, J. M. (1993). PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography, 26(2), 283–291.
Hooft, R. W., Vriend, G., Sander, C., & Abola, E. E. (1996). Errors in protein structures. Nature, 381(6580), 272.
Zhang, Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics, 9(1), 1–8. https://doi.org/10.1186/1471-2105-9-40.
Roy, A., Kucukural, A., & Zhang, Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols, 5(4), 725–738.
Zhang, J., Yang, J., Jang, R., & Zhang, Y. (2015). GPCR-I-TASSER: a hybrid approach to G protein-coupled receptor structure modeling and the application to the human genome. Structure, 23(8), 1538–1549. https://doi.org/10.1016/j.str.2015.06.007.
Shahlaei, M., & Mousavi, A. (2014). A 3D model for human melanocortin 4 receptor refined with molecular dynamics simulation. Journal of Reports in Pharmaceutical Sciences, 3(1), 42–53.
Jones, D. T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. Journal of Molecular Biology, 292(2), 195–202.
Drozdetskiy, A., Cole, C., Procter, J., & Barton, G. J. (2015). JPred4: a protein secondary structure prediction server. Nucleic Acids Research, 43(W1), W389–W394. https://doi.org/10.1093/nar/gkv332.
Geourjon, C., & Deléage, G. (1995). SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Computer Applications in the Biosciences, 11(6), 681–684. https://doi.org/10.1093/bioinformatics/11.6.681.
Mirabello, C., & Pollastri, G. (2013). Porter, PaleAle 4.0: high-accuracy prediction of protein secondary structure and relative solvent accessibility. Bioinformatics, 29(16), 2056–2058.
Kashani-Amin, E., Tabatabaei-Malazy, O., Sakhteman, A., Larijani, B., Ebrahim-Habibi, A. (2018) A systematic review on popularity, application and characteristics of protein secondary structure prediction tools. Current Drug Discovery Technologies, 15. https://doi.org/10.2174/1570163815666180227162157
Leman, J. K., Mueller, R., Karakas, M., Woetzel, N., & Meiler, J. (2013). Simultaneous prediction of protein secondary structure and transmembrane spans. Proteins: Structure, Function, and Bioinformatics, 81(7), 1127–1140. https://doi.org/10.1002/prot.24258.
Viklund, H., & Elofsson, A. (2008). OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics, 24(15), 1662–1668.
Sonnhammer E.L., Von Heijne G., Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. In: Ismb, 1998. pp 175-182
Krieger, E., Joo, K., Lee, J., Lee, J., Raman, S., Thompson, J., Tyka, M., Baker, D., & Karplus, K. (2009). Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins, 77(Suppl 9), 114–122. https://doi.org/10.1002/prot.22570.
Konagurthu, A. S., Whisstock, J. C., Stuckey, P. J., & Lesk, A. M. (2006). MUSTANG: a multiple structural alignment algorithm. Proteins: Structure, Function, and Bioinformatics, 64(3), 559–574.
Krieger, E., & Vriend, G. (2015). New ways to boost molecular dynamics simulations. Journal of Computational Chemistry, 36(13), 996–1007.
Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., & Pedersen, L. G. (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593.
Krieger, E., Nielsen, J. E., Spronk, C. A., & Vriend, G. (2006). Fast empirical pK a prediction by Ewald summation. Journal of Molecular Graphics and Modelling, 25(4), 481–486.
Krieger, E., Darden, T., Nabuurs, S. B., Finkelstein, A., & Vriend, G. (2004). Making optimal use of empirical energy functions: force‐field parameterization in crystal space. Proteins: Structure, Function, and Bioinformatics, 57(4), 678–683.
Trott, O., & Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461.
Duan, Y., Wu, C., Chowdhury, S., Lee, M. C., Xiong, G., Zhang, W., Yang, R., Cieplak, P., Luo, R., & Lee, T. (2003). A point‐charge force field for molecular mechanics simulations of proteins based on condensed‐phase quantum mechanical calculations. Journal of Computational Chemistry, 24(16), 1999–2012.
Brenke, R., Kozakov, D., Chuang, G.-Y., Beglov, D., Hall, D., Landon, M. R., Mattos, C., & Vajda, S. (2009). Fragment-based identification of druggable ‘hot spots’ of proteins using Fourier domain correlation techniques. Bioinformatics, 25(5), 621–627.
Kozakov, D., Hall, D. R., Chuang, G.-Y., Cencic, R., Brenke, R., Grove, L. E., Beglov, D., Pelletier, J., Whitty, A., & Vajda, S. (2011). Structural conservation of druggable hot spots in protein–protein interfaces. Proceedings of the National Academy of Sciences, 108(33), 13528–13533.
Ngan, C.H., Bohnuud, T., Mottarella, S.E., Beglov, D., Villar, E.A., Hall, D.R., Kozakov, D., Vajda, S. (2012) FTMAP: extended protein mapping with user-selected probe molecules. Nucleic Acids Research, 40,(Web Server issue):W271–W275. https://doi.org/10.1093/nar/gks441
Kozakov, D., Grove, L. E., Hall, D. R., Bohnuud, T., Mottarella, S. E., Luo, L., Xia, B., Beglov, D., & Vajda, S. (2015). The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nature Protocols, 10(5), 733.
Wu, H., Wang, C., Gregory, K. J., Han, G. W., Cho, H. P., Xia, Y., Niswender, C. M., Katritch, V., Meiler, J., & Cherezov, V. (2014). Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science, 344(6179), 58–64.
Acknowledgements
We thank the Non-Communicable Diseases Research Center of Endocrinology and Metabolism Population Sciences Institute and Dr. Latifeh Navidpour for allocating computational resources to help this project.
Funding
This research has been supported by the Endocrinology and Metabolism Research Institute of Tehran University of Medical Sciences.
Author Contributions
This report contains part of the results obtained from E.K.-A. Ph.D. thesis project who has done model constructions, validations, calculations and analyses, and written the manuscript text draft. A.E-H. was supervisor of the thesis, developed the initial idea and supervised the research project from data gathering to critical paper review. A.S. was advisor of the thesis and has done technical supervision of the model construction, validation, and analyses and critical paper review. B.L. was advisor of the thesis and has contributed in the idea developing and supervising project progress as an endocrine and metabolism research study. All authors have reviewed the final script and commented if necessary.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Kashani-Amin, E., Sakhteman, A., Larijani, B. et al. Introducing a New Model of Sweet Taste Receptor, a Class C G-protein Coupled Receptor (C GPCR). Cell Biochem Biophys 77, 227–243 (2019). https://doi.org/10.1007/s12013-019-00872-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-019-00872-7