Abstract
Purpose
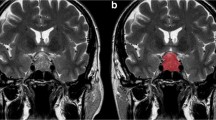
Pituitary adenomas are among the most frequent intracranial tumors. They may exhibit clinically aggressive behavior, with recurrent disease and resistance to multimodal therapy. The ki-67 labeling index represents a proliferative marker which correlates with pituitary adenoma aggressiveness. Aim of our study was to assess the accuracy of machine learning analysis of texture-derived parameters from pituitary adenomas preoperative MRI for the prediction of ki-67 proliferation index class.
Methods
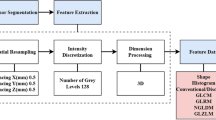
A total of 89 patients who underwent an endoscopic endonasal procedure for pituitary adenoma removal with available ki-67 labeling index were included. From T2w MR images, 1128 quantitative imaging features were extracted. To select the most informative features, different supervised feature selection methods were employed. Subsequently, a k-nearest neighbors (k-NN) classifier was employed to predict macroadenoma high or low proliferation index. Algorithm validation was performed with a train-test approach.
Results
Of the 12 subsets derived from feature selection, the best performing one was constituted by the 4 highest correlating parameters at Pearson’s test. These all showed very good (ICC ≥ 0.85) inter-observer reproducibility. The overall accuracy of the k-NN in the test group was of 91.67% (33/36) of correctly classified patients.
Conclusions
Machine learning analysis of texture-derived parameters from preoperative T2 MRI has proven to be effective for the prediction of pituitary macroadenomas ki-67 proliferation index class. This might aid the surgical strategy making a more accurate preoperative lesion classification and allow for a more focused and cost-effective follow-up and long-term management.





Similar content being viewed by others
References
Daly AF, Rixhon M, Adam C, Dempegioti A, Tichomirowa MA, Beckers A (2006) High prevalence of pituitary adenomas: a cross-sectional study in the province of Liège, Belgium. J Clin Endocrinol Metab 91:4769–4775. https://doi.org/10.1210/jc.2006-1668
Trouillas J, Roy P, Sturm N et al (2013) A new prognostic clinicopathological classification of pituitary adenomas: a multicentric case-control study of 410 patients with 8 years post-operative follow-up. Acta Neuropathol 126:123–135. https://doi.org/10.1007/s00401-013-1084-y
Paek K-I, Kim S-H, Song S-H, Choi SW, Koh HS, Youm JY, Kim Y (2005) Clinical significance of Ki-67 labeling index in pituitary macroadenoma. J Korean Med Sci 20:489. https://doi.org/10.3346/jkms.2005.20.3.489
Chatzellis E, Alexandraki KI, Androulakis II, Kaltsas G (2015) Aggressive pituitary tumors. Neuroendocrinology 101:87–104. https://doi.org/10.1159/000371806
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577. https://doi.org/10.1148/radiol.2015151169
Senders JT, Zaki MM, Karhade AV, Chang B, Gormley WB, Broekman ML, Smith TR, Arnaout O (2018) An introduction and overview of machine learning in neurosurgical care. Acta Neurochir 160:29–38. https://doi.org/10.1007/s00701-017-3385-8
Stanzione A, Cuocolo R, Cocozza S, Romeo V, Persico F, Fusco F, Longo N, Brunetti A, Imbriaco M (2019) Detection of extraprostatic extension of cancer on biparametric MRI combining texture analysis and machine learning: preliminary results. Acad Radiol:1–7. https://doi.org/10.1016/j.acra.2018.12.025
Romeo V, Maurea S, Cuocolo R, Petretta M, Mainenti PP, Verde F, Coppola M, Dell’Aversana S, Brunetti A (2018) Characterization of adrenal lesions on unenhanced MRI using texture analysis: a machine-learning approach. J Magn Reson Imaging 48:198–204. https://doi.org/10.1002/jmri.25954
Kadir T, Gleeson F (2018) Lung cancer prediction using machine learning and advanced imaging techniques. Transl Lung Cancer Res 7:304–312. https://doi.org/10.21037/tlcr.2018.05.15
Fan M, Cheng H, Zhang P, Gao X, Zhang J, Shao G, Li L (2018) DCE-MRI texture analysis with tumor subregion partitioning for predicting Ki-67 status of estrogen receptor-positive breast cancers. J Magn Reson Imaging 48:237–247. https://doi.org/10.1002/jmri.25921
Imbriaco M, Cuocolo R (2018) Does texture analysis of MR images of breast tumors help predict response to treatment? Radiology 286:421–423. https://doi.org/10.1148/radiol.2017172454
Liu S, Zheng H, Zhang Y, Chen L, Guan W, Guan Y, Ge Y, He J, Zhou Z (2018) Whole-volume apparent diffusion coefficient-based entropy parameters for assessment of gastric cancer aggressiveness. J Magn Reson Imaging 47:168–175. https://doi.org/10.1002/jmri.25752
Cuocolo R, Stanzione A, Ponsiglione A, Romeo V, Verde F, Creta M, la Rocca R, Longo N, Pace L, Imbriaco M (2019) Clinically significant prostate cancer detection on MRI: a radiomic shape features study. Eur J Radiol 116:144–149. https://doi.org/10.1016/j.ejrad.2019.05.006
Romeo V, Ricciardi C, Cuocolo R, Stanzione A, Verde F, Sarno L, Improta G, Mainenti PP, D’Armiento M, Brunetti A, Maurea S (2019) Machine learning analysis of MRI-derived texture features to predict placenta accreta spectrum in patients with placenta previa. Magn Reson Imaging. https://doi.org/10.1016/j.mri.2019.05.017
Cappabianca P, Cavallo LM, Solari D, Stagno V, Esposito F, de Angelis M (2014) Endoscopic endonasal surgery for pituitary adenomas. World Neurosurg 82:S3–S11. https://doi.org/10.1016/j.wneu.2014.07.019
de Notaris M, Solari D, Cavallo LM, D’Enza AI, Enseñat J, Berenguer J, Ferrer E, Prats-Galino A, Cappabianca P (2012) The “suprasellar notch,” or the tuberculum sellae as seen from below: definition, features, and clinical implications from an endoscopic endonasal perspective. J Neurosurg 116:622–629. https://doi.org/10.3171/2011.11.JNS111162
Kassam A, Snyderman CH, Mintz A, et al (2005) Expanded endonasal approach: the rostrocaudal axis. Part I. Crista galli to the sella turcica. Neurosurg Focus
Solari D, Cavallo LM, Cappabianca P (2014) Surgical approach to pituitary tumors. Handb Clin Neurol 124:291–301. https://doi.org/10.1016/B978-0-444-59602-4.00019-8
Cappabianca P, Cavallo LM, Esposito F, de Divitiis O, Messina A, de Divitiis E (2008) Extended endoscopic endonasal approach to the midline skull base: the evolving role of transsphenoidal surgery. In: Pickard JD, Akalan N, Di Rocco C et al (eds) Advances and technical standards in neurosurgery. Springer Vienna, Vienna, pp 151–199
Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31:1116–1128. https://doi.org/10.1016/j.neuroimage.2006.01.015
van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts HJWL (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107. https://doi.org/10.1158/0008-5472.CAN-17-0339
Leijenaar RTH, Nalbantov G, Carvalho S, van Elmpt WJC, Troost EGC, Boellaard R, Aerts HJWL, Gillies RJ, Lambin P (2015) The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep 5:11075. https://doi.org/10.1038/srep11075
Haralick RM, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern SMC-3:610–621. https://doi.org/10.1109/TSMC.1973.4309314
THIBAULT G, FERTIL B, NAVARRO C, PEREIRA S, CAU P, LEVY N, SEQUEIRA J, MARI JL (2013) Shape and texture indexes application to cell nuclei classification. Int J Pattern Recognit Artif Intell 27:1357002. https://doi.org/10.1142/S0218001413570024
Galloway MM (1975) Texture analysis using gray level run lengths. Comput Graph Image Process 4:172–179. https://doi.org/10.1016/S0146-664X(75)80008-6
Amadasun M, King R (1989) Textural features corresponding to textural properties. IEEE Trans Syst Man Cybern 19:1264–1274. https://doi.org/10.1109/21.44046
Sun C, Wee WG (1983) Neighboring gray level dependence matrix for texture classification. Comput Vision, Graph Image Process 23:341–352. https://doi.org/10.1016/0734-189X(83)90032-4
Cai J, Luo J, Wang S, Yang S (2018) Feature selection in machine learning: a new perspective. Neurocomputing 300:70–79. https://doi.org/10.1016/j.neucom.2017.11.077
Eibe F, Hall MA, Witten IH (2016) The WEKA Workbench. Online Appendix for “Data Mining: Practical Machine Learning Tools and Techniques,” Fourth Edi. Morgan Kaufmann
Juntu J, Sijbers J, De Backer S et al (2010) Machine learning study of several classifiers trained with texture analysis features to differentiate benign from malignant soft-tissue tumors in T1-MRI images. J Magn Reson Imaging 31:680–689. https://doi.org/10.1002/jmri.22095
Siva Kumar N (1996) An affinity method for the purification of mannose 6-phosphate receptor proteins (MPR 215) from rat tissues and goat liver. J Biochem Biophys Methods 31:181–184. https://doi.org/10.1016/0165-022X(95)00026-N
Aha DW, Kibler D, Albert MK (1991) Instance-based learning algorithms. Mach Learn 6:37–66. https://doi.org/10.1023/A:1022689900470
Thapar K, Kovacs K, Scheithauer BW, Stefaneanu L, Horvath E, Peter J. P, Murray D, Laws ER Jr (1996) Proliferative activity and invasiveness among pituitary adenomas and carcinomas: an analysis using the MIB-1 antibody. Neurosurgery 38:99–107. https://doi.org/10.1097/00006123-199601000-00024
Brennan P, Silman A (1992) Statistical methods for assessing observer variability in clinical measures. BMJ 304:1491–1494. https://doi.org/10.1136/bmj.304.6840.1491
Del Basso De Caro M, Solari D, Pagliuca F et al (2017) Atypical pituitary adenomas: clinical characteristics and role of ki-67 and p53 in prognostic and therapeutic evaluation. A series of 50 patients. Neurosurg Rev 40:105–114. https://doi.org/10.1007/s10143-016-0740-9
Vasiljevic A, Jouanneau E, Trouillas J, Raverot G (2016) Clinicopathological prognostic and theranostic markers in pituitary tumors. Minerva Endocrinol 41:377–389
Mete O, Lopes MB (2017) Overview of the 2017 WHO classification of pituitary tumors. Endocr Pathol 28:228–243. https://doi.org/10.1007/s12022-017-9498-z
Inoshita N, Nishioka H (2018) The 2017 WHO classification of pituitary adenoma: overview and comments. Brain Tumor Pathol 35:51–56. https://doi.org/10.1007/s10014-018-0314-3
Hollon TC, Parikh A, Pandian B, Tarpeh J, Orringer DA, Barkan AL, McKean EL, Sullivan SE (2018) A machine learning approach to predict early outcomes after pituitary adenoma surgery. Neurosurg Focus 45:1–9. https://doi.org/10.3171/2018.8.FOCUS18268
Staartjes VE, Serra C, Muscas G, et al (2018) Utility of deep neural networks in predicting gross-total resection after transsphenoidal surgery for pituitary adenoma: a pilot study 45:1–7 . doi: https://doi.org/10.3171/2018.8.FOCUS18243
Zhang S, Song G, Zang Y, Jia J, Wang C, Li C, Tian J, Dong D, Zhang Y (2018) Non-invasive radiomics approach potentially predicts non-functioning pituitary adenomas subtypes before surgery. Eur Radiol 28:3692–3701. https://doi.org/10.1007/s00330-017-5180-6
Niu J, Tian J, Zhou W et al (2018) Preoperative prediction of cavernous sinus invasion by pituitary adenomas using a radiomics method based on magnetic resonance images. Eur Radiol 29:1625–1634. https://doi.org/10.1007/s00330-018-5725-3
Tamrazi B, Pekmezci M, Aboian M, Tihan T, Glastonbury CM (2017) Apparent diffusion coefficient and pituitary macroadenomas: pre-operative assessment of tumor atypia. Pituitary 20:195–200. https://doi.org/10.1007/s11102-016-0759-5
Hua J, Xiong Z, Lowey J, Suh E, Dougherty ER (2005) Optimal number of features as a function of sample size for various classification rules. Bioinformatics 21:1509–1515. https://doi.org/10.1093/bioinformatics/bti171
Zaharchuk G, Gong E, Wintermark M, Rubin D, Langlotz CP (2018) Deep learning in neuroradiology. Am J Neuroradiol 39:1776–1784. https://doi.org/10.3174/ajnr.A5543
Sanei Taheri M, Kimia F, Mehrnahad M, Saligheh Rad H, Haghighatkhah H, Moradi A, Kazerooni AF, Alviri M, Absalan A (2019) Accuracy of diffusion-weighted imaging-magnetic resonance in differentiating functional from non-functional pituitary macro-adenoma and classification of tumor consistency. Neuroradiol J 32:74–85. https://doi.org/10.1177/1971400918809825
Wei L, Lin S-A, Fan K et al (2015) Relationship between pituitary adenoma texture and collagen content revealed by comparative study of MRI and pathology analysis. Int J Clin Exp Med 8:12898–12905
Zeynalova A, Kocak B, Durmaz ES, Comunoglu N, Ozcan K, Ozcan G, Turk O, Tanriover N, Kocer N, Kizilkilic O, Islak C (2019) Preoperative evaluation of tumour consistency in pituitary macroadenomas: a machine learning-based histogram analysis on conventional T2-weighted MRI. Neuroradiology 61:767–774. https://doi.org/10.1007/s00234-019-02211-2
Heck A, Emblem KE, Casar-Borota O, Bollerslev J, Ringstad G (2016) Quantitative analyses of T2-weighted MRI as a potential marker for response to somatostatin analogs in newly diagnosed acromegaly. Endocrine 52:333–343. https://doi.org/10.1007/s12020-015-0766-8
Kocak B, Durmaz ES, Kadioglu P, Polat Korkmaz O, Comunoglu N, Tanriover N, Kocer N, Islak C, Kizilkilic O (2019) Predicting response to somatostatin analogues in acromegaly: machine learning-based high-dimensional quantitative texture analysis on T2-weighted MRI. Eur Radiol 29:2731–2739. https://doi.org/10.1007/s00330-018-5876-2
Funding
No funding was received for this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in the studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Requirement for informed consent was waived by the local IRB.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ugga, L., Cuocolo, R., Solari, D. et al. Prediction of high proliferative index in pituitary macroadenomas using MRI-based radiomics and machine learning. Neuroradiology 61, 1365–1373 (2019). https://doi.org/10.1007/s00234-019-02266-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00234-019-02266-1