Abstract
Background
Various kinds of transposable elements (TEs) constitute high proportions of eukaryotic genomes. Although most of these TEs are not actively mobile, genome stress can induce mobilization of dormant TEs. Transgenic plants undergo tissue culture and subsequent whole-plant regeneration, which can cause genomic stress and in turn induce mobilization of inactive TEs.
Objectives
To investigate the activation of transposable elements on the genome wide of the GM plant.
Methods
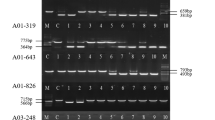
Transposon activities were analyzed in three transgenic rice plants carrying the insect resistance gene Cry1Ac and an herbicide resistance gene by the transposon display technique. These three transgenic plants were derived from a leading Korean rice variety, Illmi.
Results
We detected seven mobile activities in the mPing element, which is a MITE family transposon. The identity of the novel fragments in the gel display was confirmed by checking TAA target site duplication via sequence analysis. The genomic integration sites were all on different chromosomes, and the integrations were specific to either one or two T1 transgenic lines, except for one common integration on chromosome 4. One integration was in the 5′-UTR of the Glycerol-3-phosphate acyltransferase 8 gene, two integrations were in introns of expressed genes, and the other four integrations were in intergenic regions.
Conclusion
Thus, novel mobilization of dormant TEs occurs in transgenic plants, which must be considered in the generation of genetically modified crops (GM crops).




Similar content being viewed by others
References
Adkins S, Kunanuvatchaidach R, Godwin ID (1995) Somaclonal variation in rice-2 drought tolerance and other agronomic characters. Austral J Bot 43:201–209
Bertin P, Bouharmont J, Kinet NM (1997) Somaclonal variation and improvement of chilling tolerance in rice—changes in chilling induced chlorophyll fluorescence. Crop Sci 37:1727–1735
Bureau TE, Wessler SR (1992) Tourist: a large family of small invert-repeat transposable elements frequently associated maize genes. Plant Cell 4:1283–1294
Bureau TE, Wessler SR (1994a) Mobile inverted-repeat element Tourist are associated with the genes in many cereal grasses. PNAS 91:1411–1415
Bureau TE, Wessler SR (1994b) Stowaway: a new family of inverted-repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell 6:907–916
Choi JY, Roy NS, Park KC, Kim NS (2016) Comparison of molecular genetic utilities of TD, AFLP, and MSAP among the accessions of japonica, indica, and Tongil of Oryza sativa L. Genes Genom 38:819–830
Heinz D, Mee GW (1971) Morphologic, cytogenetic, and enzymatic variation in Saccharum species hybrid clones derived from callus tissue. Am J Bot 58:257–262
Hirochika H (1993) Activation of tobacco retrotranposons during tissue culture. EMBO J 12:2521–2528
Hirochika H (2001) Contribution of the Tos17 retrotransposon to rice functional genomics. Curr Opin Plant Biol 4:118–122
Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M (1996) Retrotransposons of rice involved in mutations induced by tissue culture. PNAS 93:7783–7788
Jiang N, Bao Z, Zhang X, Hirochioka H, Eddy SR, McCouch SR, Wessler SR (2003) An active DNA transposon family in rice. Nature 421:163–167
Jiang C, Mithani A, Gan X, Belfield EJ, Klinger JP, Zhu JK, Ragoussis J, Mott R, Harberd NP (2011) Regenerant Arabidopsis lineages display a distinctive genome-wide spectrum of mutation conferring variant phenotypes. Curr Biol 21:1385–1390
Joshi RK, Rao GJN (2009) Somaclonal variation in submergence tolerance rice cultivars and induced diversity evaluation by PCR markers. Int J Genet Mol Biol 1:010–088
Karp A (1995) Somaclonal variation as a tool for crop improvement. Euphytica 85:295–302
Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu JZ, Zhou SG et al (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. https://doi.org/10.1186/1939-8433-6-4
Kikuchi K, Terauchi K, Wada M, Hirano HY (2003) The plant MITE mPing is mobilized in anther culture. Nature 421:167–170
Korswagen HC, Durbin RM, Smits MT, Plsterk RHA (1996) Transposon TC1-derived, sequence-tagged sites in Caenohabiditis elegans as markers for genetic mapping. PNAS 93:14680–14685
Kwon SJ, Lee JK, Hong SW, Park YJ, McNally KL, Kim NS (2006) Genetic diversity and phylogenetic relationship in AA Oryza species as revealed by Rim2/Hippa CACTA transposon display. Genes Genet Syst 81:93–101
Larkin PJ, Scowcroft WR (1981) Somaclonal variation—a novel source of variability from cell cultures for plant improvement. Theor Appl Genet 20:197–210
Le QH, Bureau T (2004) Prediction and quality assessment of transposon insertion display data. Biotechnique 36:222–228
Lee SI, Kim NS (2014) Transposable elements and genome size variation in plants. Genomics Inform 12:87–97
McClintock B (1984) The significance of responses of the genome to challenge. Science 226:792–801
Miyao A, Nakagome M, Ohnuma T, Yamagata H, Kanamori H, Katayose Y, Takahashi A, Matsumoto T, Hirochika H (2012) Molecular spectrum of somaclonal variation in regenerated rice revealed by whole-genome sequencing. Plant Cell Physiol 53:256–264
Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Pkumoto Y, Tanisaka T, Wessler SR (2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461:1130–1134
Nakazaki T, Okumoto Y, Horibata A, Yamahira S, Teraishi M, Nishida H, Inoue H, Taisaka T (2003) Mobilization of a transposon in the rice genome. Nature 421:170–172
Neelakandan AK, Wang K (2012) Recent progress in the understanding of tissue culture-induced genome level changes in plants and potential applications. Plant Cell Rep 31:597–620
Park D, Park SH, Ban YW, Kim YS, Park KC, Kim NS, Kim JK, Choi IY (2017) A bioinformatics approach for identifying transgene insertion sites using whole genome sequencing data. BMC Biotechnol 17:67. https://doi.org/10.1186/s12896-017-0386-x
Park D, Park SH, Kim YS, Choi BS, Kim JK, Kim NS, Choi IY (2019) NGS sequencing reveals that many of the genetic variations in transgenic rice plants match the variations found in natural rice population. Genes Genom 41:213–222
Roy NS, Choi JY, Lee SI, Kim NS (2015) Marker utility of transposable elements for plant genetics, breeding, and ecology: a review. Genes Genom 37:141–151
Sabot F, Picault N, El-Baidouri M, Llauro C, Chaparro C, Peigu B, Roulin A, Guiderdoni E, Delabastide M, McCombie R, Panaud O (2011) Transpositional landscape of the rice genome revealed by paired-end mapping of high-throughput re-sequencing data. Plant J 66:241–246
Shan X, Liu Z, Dong Z, Wang Y, Chen Y, Lin X, Long L, Han F, Dong Y, Liu B (2005) Mobilization of the active MITE transposons mPing and Pong in rice by introgression from wild rice (Zizania latifolia Griseb.). Mol Biol Evol 22:976–990
Smith AM, Hansey CN, Kaeppler SM (2012) TCUP: A novel hAT transposon active in maize tissue culture. Front Plant Sci. https://doi.org/10.3389/fpls.2012.00006
Stein JC, Yu Y, Copetti D, Zwickl DJ, Zhang L, Zhang C, Chougule K, Gao D, Iwata A, Goicoechea JL et al (2018) Genomes of 13 domesticated and wild rice highlight genetic conservation, turnover and innovation across the genus Oryza. Nat Genet 50:285–286
Teraishi M, Okumoto Y, Hirochika H, Horibata A, Yamagata H, Tanisaka T (1999) Identification of mutable slender glume gene in rice (Oryza sativa L.). Mol Gen Genet 261:487–494
Zhang P, Li J, Li X, Liu X, Zhao X, Lu Y (2011) Population structure and genetic diversity in rice core collection (Oryza sativa L.) investigated with SR markers. PLoS One 6(12):e27565
Zhang D, Wang Z, Wang N, Gao Y, Liu Y, Wu Y, Bai Y, Zhang Z, Lin X, Dong Y et al (2014) Tissue culture-induced heritable genome variation in rice, and their phenotypic implications. PLoS One 9:e96879. https://doi.org/10.1371/journal.pone.0096879
Acknowledgements
This work was supported by the Next-Generation BioGreen21 Program (PJ01131301), Rural Development Administration of the Korean government.
Author information
Authors and Affiliations
Contributions
IYC designed and oversaw the project. NSK wrote the manuscript. DP was a contributor to experiments and drafted the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing of interests.
Ethical approval
This study does not contain any performing with human and animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Park, D., Choi, IY. & Kim, NS. Detection of mPing mobilization in transgenic rice plants. Genes Genom 42, 47–54 (2020). https://doi.org/10.1007/s13258-019-00877-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-019-00877-9