Abstract
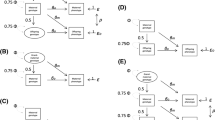
There is increasing interest within the genetics community in estimating the relative contribution of parental genetic effects on offspring phenotypes. Here we describe the user-friendly M-GCTA software package used to estimate the proportion of phenotypic variance explained by maternal (or alternatively paternal) and offspring genotypes on offspring phenotypes. The tool requires large studies where genome-wide genotype data are available on mother- (or alternatively father-) offspring pairs. The software includes several options for data cleaning and quality control, including the ability to detect and automatically remove cryptically related pairs of individuals. It also allows users to construct genetic relationship matrices indexing genetic similarity across the genome between parents and offspring, enabling the estimation of variance explained by maternal (or alternatively paternal) and offspring genetic effects. We evaluated the performance of the software using a range of data simulations and estimated the computing time and memory requirements. We demonstrate the use of M-GCTA on previously analyzed birth weight data from two large population based birth cohorts, the Avon Longitudinal Study of Parents and Children (ALSPAC) and the Norwegian Mother and Child Cohort Study (MoBa). We show how genetic variation in birth weight is predominantly explained by fetal genetic rather than maternal genetic sources of variation.



Similar content being viewed by others
References
Beaumont RN, Warrington NM, Cavadino A, Tyrrell J, Nodzenski M, Horikoshi M, Geller F, Myhre R, Richmond RC, Paternoster L, Bradfield JP, Kreiner-Møller E, Huikari V, Metrustry S, Lunetta KL, Painter JN, Hottenga JJ, Allard C, Barton SJ, Espinosa A, Marsh JA, Potter C, Zhang G, Ang W, Berry DJ, Bouchard L, Das S, Early Growth Genetics (EGG) Consortium, Hakonarson H, Heikkinen J, Helgeland Ø, Hocher B, Hofman A, Inskip HM, Jones SE, Kogevinas M, Lind PA, Marullo L, Medland SE, Murray A, Murray JC, Njølstad PR, Nohr EA, Reichetzeder C, Ring SM, Ruth KS, Santa-Marina L, Scholtens DM, Sebert S, Sengpiel V, Tuke MA, Vaudel M, Weedon MN, Willemsen G, Wood AR, Yaghootkar H, Muglia LJ, Bartels M, Relton CL, Pennell CE, Chatzi L, Estivill X, Holloway JW, Boomsma DI, Montgomery GW, Murabito JM, Spector TD, Power C, Järvelin MR, Bisgaard H, Grant SFA, Sørensen TIA, Jaddoe VW, Jacobsson B, Melbye M, McCarthy MI, Hattersley AT, Hayes MG, Frayling TM, Hivert MF, Felix JF, Hyppönen E, Lowe WL Jr, Evans DM, Lawlor DA, Feenstra B, Freathy RM (2018) Genome-wide association study of offspring birth weight in 86,577 women identifies five novel loci and highlights maternal genetic effects that are independent of fetal genetics. Hum Mol Genet 27:742–756
Bernardo J (1996) Maternal effects in animal ecology. Am Zool 36:83–105
Boker S, Neale M, Maes H, Wilde M, Spiegel M, Brick T, Spies J, Estabrook R, Kenny S, Bates T, Mehta P, Fox J (2011) OpenMx: An open source extended structural equation modeling framework. Psychometrika 76:306–317
Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, Molloy L, Ness A, Ring S, Davey Smith G (2013) Cohort profile: the ‘children of the 90s’—the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol 42:111–127
Corey L, Nance W (1978) The monozygotic half-sib model: a tool for epidemiologic research. Prog Clin Biol Res 24A:201–209
Cuellar Partida G, Laurin C, Ring SM, Gaunt TR, McRae A, Visscher PM, Montgomery G, Martin NG, Hemani G, Suderman M, Relton CL, Davey Smith G, Evans DM (2018) Genome-wide survey of parent-of-origin effects on DNA methylation identifies candidate imprinted loci in humans. Hum Mol Genet 27:2927–2939
Curley JP, Mashoodh R, Champagne FA (2011) Epigenetics and the origins of paternal effects. Horm Behav 59:306–314
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Eaves LJ, Pourcain BS, Smith GD, York TP, Evans DM (2014) Resolving the effects of maternal and offspring genotype on dyadic outcomes in genome wide complex trait analysis (“M-GCTA”). Behav Genet 44:445–455
Evans DM, Moen G-H, Hwang D, Lawlor DA, Warrington NM (2019) Elucidating the role of maternal environmental exposures on offspring health and disease using two-sample mendelian randomization. Int J Epidemiol 48:861–875
Evans DM, Visscher PM, Wray NR (2009) Harnessing the information contained within genome-wide association studies to improve individual prediction of complex disease risk. Hum Mol Genet 18:3525–3531
Evans DM, Zhu G, Dy V, Heath AC, Madden PA, Kemp JP, McMahon G, St Pourcain B, Timpson NJ, Golding J, Lawlor DA, Steer C, Montgomery GW, Martin NG, Smith GD, Whitfield JB (2013) Genome-wide association study identifies loci affecting blood copper, selenium and zinc. Hum Mol Genet 22:3998–4006
Falconer DS, Mackay TFC (1996) Introduction to quantitative genetics. Longman, London
Fatemifar G, Hoggart CJ, Paternoster L, Kemp JP, Prokopenko I, Horikoshi M, Wright VJ, Tobias JH, Richmond S, Zhurov AI, Toma AM, Pouta A, Taanila A, Sipila K, Lähdesmäki R, Pillas D, Geller F, Feenstra B, Melbye M, Nohr EA, Ring SM, St Pourcain B, Timpson NJ, Davey Smith G, Jarvelin MR, Evans DM (2013) Genome-wide association study of primary tooth eruption identifies pleiotropic loci associated with height and craniofacial distances. Hum Mol Genet 22:3807–3817
Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Davey Smith G, Henderson J, Macleod J, Molloy L, Ness A (2012) Cohort profile: the avon longitudinal study of parents and children: ALSPAC mothers cohort. Int J Epidemiol 42:97–110
Guilmatre A, Sharp A (2012) Parent of origin effects. Clin Genet 81:201–209
Hewitt JK (2015) Announcement of the Fulker Award for a Paper Published in Behavior Genetics, Volume 44, 2014. Behav Genet 45:699
Horikoshi M, Beaumont RN, Day FR, Warrington NM, Kooijman MN, Fernandez-Tajes J, Feenstra B, van Zuydam NR, Gaulton KJ, Grarup N, Bradfield JP, Strachan DP, Li-Gao R, Ahluwalia TS, Kreiner-Møller E, Rueedi R, Lyytikäinen L, Cousminer DL, Wu Y, Thiering E, Wang CA, Have CT, Hottenga JJ, Vilor-Tejedor N, Joshi PK, Tai Hui Boh E, Ntalla I, Pitkänen N, Mahajan A, van Leeuwen EM, Joro R, Lagou V, Nodzenski M, Diver LA, Zondervan KT, Bustamante M, Marques-Vidal P, Mercader JM, Bennett AJ, Rahmioglu N, Nyholt DR, Ma RCW, Tam CHT, Tam WH, CHARGE Consortium Hematology Working Group, Ganesh SK, van Rooij FJA, Jones SE, Loh PR, Ruth KS, Tuke MA, Tyrrell J, Wood AR, Yaghootkar H, Scholtens DM, Paternoster L, Prokopenko I, Kovacs P, Atalay M, Willems SM, Panoutsopoulou K, Wang X, Carstensen L, Geller F, Schraut KE, Murcia M, van Beijsterveldt CEM, Willemsen G, Appel EVR, Fonvig CE, Trier C, Tiesler CMT, Standl M, Kutalik Z, Bonas-Guarch S, Hougaard DM, Sánchez F, Torrents D, Waage J, Hollegaard MV, de Haan HG, Rosendaal FR, Medina-Gomez C, Ring SM, Hemani G, McMahon G, Robertson NR, Groves CJ, Langenberg C, Luan J, Scott RA, Zhao JH, Mentch FD, MacKenzie SM, Reynolds RM, Lowe WL Jr, Tönjes A, Stumvoll M, Lindi V, Lakka TA, van Duijn CM, Kiess W, Körner A, Sørensen TIA, Niinikoski H, Pahkala K, Raitakari OT, Zeggini E, Dedoussis GV, Teo YY, Saw SM, Melbye M, Campbell H, Wilson JF, Vrijheid M, de Geus EJCN, Boomsma DI, Kadarmideen HN, Holm JC, Hansen T, Sebert S, Hattersley AT, Beilin LJ, Newnham JP, Pennell CE, Heinrich J, Adair LS, Borja JB, Mohlke KL, Eriksson JG, Widén EE, Kähönen M, Viikari JS, Lehtimäki T, Vollenweider P, Bønnelykke K, Bisgaard H, Mook-Kanamori DO, Hofman A, Rivadeneira F, Uitterlinden AG, Pisinger C, Pedersen O, Power C, Hyppönen E, Wareham NJ, Hakonarson H, Davies E, Walker BR, Jaddoe VWV, Jarvelin MR, Grant SFA, Vaag AA, Lawlor DA, Frayling TM, Davey Smith G, Morris AP, Ong KK, Felix JF, Timpson NJ, Perry JRB, Evans DM, McCarthy MI, Freathy RM (2016) Genome-wide associations for birth weight and correlations with adult disease. Nature 538:248–252
Kong A, Thorleifsson G, Frigge ML, Vilhjalmsson BJ, Young AI, Thorgeirsson TE, Benonisdottir S, Oddsson A, Halldorsson BV, Masson G (2018) The nature of nurture: effects of parental genotypes. Science 359:424–428
Krokstad S, Langhammer A, Hveem K, Holmen T, Midthjell K, Stene T, Bratberg G, Heggland J, Holmen J (2012) Cohort profile: the HUNT study, Norway. Int J Epidemiol 42:968–977
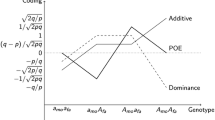
Laurin C, Cuellar-Partida G, Hemani G, Smith GD, Yang J, Evans DM (2018) Partitioning phenotypic variance due to parent-of-origin effects using genomic relatedness matrices. Behav Genet 48:67–79
Lawson HA, Cheverud JM, Wolf JB (2013) Genomic imprinting and parent-of-origin effects on complex traits. Nat Rev Genet 14:609–617
Lunde A, Melve KK, Gjessing HK, Skjærven R, Irgens LM (2007) Genetic and environmental influences on birth weight, birth length, head circumference, and gestational age by use of population-based parent-offspring data. Am J Epidemiol 165:734–741
Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer, Sunderland
Magnus P (1984a) Causes of variation in birth weight: a study of offspring of twins. Clin Genet 25:15–24
Magnus P (1984b) Further evidence for a significant effect of fetal genes on variation in birth weight. Clin Genet 26:289–296
Magnus P, Birke C, Vejrup K, Haugan A, Alsaker E, Daltveit AK, Handal M, Haugen M, Høiseth G, Knudsen GP (2016) Cohort profile update: the Norwegian mother and child cohort study (MoBa). Int J Epidemiol 45:382–388
Maher B (2008) Personal genomes: the case of the missing heritability. Nature 456:18–21
Masuda K, Osada H, Iitsuka Y, Seki K, Sekiya S (2002) Positive association of maternal G protein β3 Subunit 825T allele with reduced head circumference at birth. Pediatr Res 52:687–691
Mather K, Jinks JL (1982) Components of variation. In: Mather K, Jinks JL (eds) Biometrical genetics. Springer, Boston, pp 135–175
Meyer K (1992) Variance components due to direct and maternal effects for growth traits of Australian beef cattle. Livest Prod Sci 31:179–204
Moen G-H, Hemani G, Warrington NM, Evans DM (2019) Calculating power to detect maternal and offspring genetic effects in genetic association studies. Behav Genet 49:327–339
Morison IM, Ramsay JP, Spencer HG (2005) A census of mammalian imprinting. Trends Genet 21:457–465
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575
R Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org
Rando OJ (2012) Daddy issues: paternal effects on phenotype. Cell 151:702–708
Rice F, Thapar A (2010) Estimating the relative contributions of maternal genetic, paternal genetic and intrauterine factors to offspring birth weight and head circumference. Early Hum Dev 86:425–432
Richmond RC, Al-Amin A, Smith GD, Relton CL (2014) Approaches for drawing causal inferences from epidemiological birth cohorts: a review. Early Hum Dev 90:769–780
Schwarzer G (2007) meta: An R package for meta-analysis. R News 7:40–45
Smith GD (2008) Assessing intrauterine influences on offspring health outcomes: can epidemiological studies yield robust findings? Basic Clin Pharmacol Toxicol 102:245–256
Smith GD (2012) Negative control exposures in epidemiological studies. Epidemiology 23:350–351
Speed D, Cai N, Johnson MR, Nejentsev S, Balding DJ, Consortium U (2017) Reevaluation of SNP heritability in complex human traits. Nat Genet 49:986–992
Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M (2015) UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12(3):e1001779
Taal HR, St Pourcain B, Thiering E, Das S, Mook-Kanamori DO, Warrington NM, Kaakinen M, Kreiner-Møller E, Bradfield JP, Freathy RM, Geller F, Guxens M, Cousminer DL, Kerkhof M, Timpson NJ, Ikram MA, Beilin LJ, Bønnelykke K, Buxton JL, Charoen P, Chawes BLK, Eriksson J, Evans DM, Hofman A, Kemp JP, Kim CE, Klopp N, Lahti J, Lye SJ, McMahon G, Mentch FD, Müller M, O'Reilly PF, Prokopenko I, Rivadeneira F, Steegers EAP, Sunyer J, Tiesler C, Yaghootkar H, Cohorts for Heart and Aging Research in Genetic Epidemiology (CHARGE) Consortium, Breteler MMB, Debette S, Fornage M, Gudnason V, Launer LJ, van der Lugt A, Mosley TH, Seshadri S, Smith AV, Vernooij MW, Early Genetics & Lifecourse Epidemiology (EAGLE) consortium, Blakemore AI, Chiavacci RM, Feenstra B, Fernandez-Benet J, Grant SFA, Hartikainen AL, van der Heijden AJ, Iñiguez C, Lathrop M, McArdle WL, Mølgaard A, Newnham JP, Palmer LJ, Palotie A, Pouta A, Ring SM, Sovio U, Standl M, Uitterlinden AG, Wichmann HE, Vissing NH, DeCarli C, van Duijn CM, McCarthy MI, Koppelman GH, Estivill X, Hattersley AT, Melbye M, Bisgaard H, Pennell CE, Widen E, Hakonarson H, Smith GD, Heinrich J, Jarvelin MR, Early Growth Genetics (EGG) Consortium, Jaddoe VWV (2012) Common variants at 12q15 and 12q24 are associated with infant head circumference. Nat Genet 44:532–538
Timpson NJ, Greenwood CM, Soranzo N, Lawson DJ, Richards JB (2018) Heritable contributions versus genetic architecture. Nat Rev Genet 19:185
van der Valk RJ, Kreiner-Møller E, Kooijman MN, Guxens M, Stergiakouli E, Sääf A, Bradfield JP, Geller F, Hayes MG, Cousminer DL (2014) A novel common variant in DCST2 is associated with length in early life and height in adulthood. Hum Mol Genet 24:1155–1168
Warrington NM, Beaumont RN, Horikoshi M, Day FR, Helgeland Ø, Laurin C, Bacelis J, Peng S, Hao K, Feenstra B, Wood AR, Mahajan A, Tyrrell J, Robertson NR, Rayner W, Qiao Z, Moen GH, Vaudel M, Marsit CJ, Chen J, Nodzenski M, Schnurr TM, Zafarmand MH, Bradfield JP, Grarup N, Kooijman MN, Li-Gao R, Geller F, Ahluwalia TS, Paternoster L, Rueedi R, Huikari V, Hottenga JJ, Lyytikäinen LP, Cavadino A, Metrustry S, Cousminer DL, Wu Y, Thiering E, Wang CA, Have CT, Vilor-Tejedor N, Joshi PK, Painter JN, Ntalla I, Myhre R, Pitkänen N, van Leeuwen EM, Joro R, Lagou V, Richmond RC, Espinosa A, Barton SJ, Inskip HM, Holloway JW, Santa-Marina L, Estivill X, Ang W, Marsh JA, Reichetzeder C, Marullo L, Hocher B, Lunetta KL, Murabito JM, Relton CL, Kogevinas M, Chatzi L, Allard C, Bouchard L, Hivert MF, Zhang G, Muglia LJ, Heikkinen J, Early Growth Genetics (EGG) Consortium, Morgen CS, van Kampen AHC, van Schaik BDC, Mentch FD, Langenberg C, Luan J, Scott RA, Hua Zhao JH, Hemani G, Ring SM, Bennett AJ, Gaulton KJ, Fernandez-Tajes J, van Zuydam NR, Medina-Gomez C, de Haan HG, Rosendaal FR, Kutalik Z, Marques-Vidal P, Das S, Willemsen G, Mbarek H, Müller-Nurasyid M, Standl M, Appel EVR, Fonvig CE, Trier C, van Beijsterveldt CEM, Murcia M, Bustamante M, Bonas-Guarch S, Hougaard DM, Mercader JM, Linneberg A, Schraut KE, Lind PA, Medland SE, Shields BM, Knight BA, Chai JF, Panoutsopoulou K, Bartels M, Sánchez F, Stokholm J, Torrents D, Vinding RK, Willems SM, Atalay M, Chawes BL, Kovacs P, Prokopenko I, Tuke MA, Yaghootkar H, Ruth KS, Jones SE, Loh PR, Murray A, Weedon MN, Tönjes A, Stumvoll M, Michaelsen KF, Eloranta AM, Lakka TA, van Duijn CM, Kiess W, Körner A, Niinikoski H, Pahkala K, Raitakari OT, Jacobsson B, Zeggini E, Dedoussis GV, Teo YY, Saw SM, Montgomery GW, Campbell H, Wilson JF, Vrijkotte TGM, Vrijheid M, de Geus EJCN, Geoffrey Hayes M, Kadarmideen HN, Holm JC, Beilin LJ, Pennell CE, Heinrich J, Adair LS, Borja JB, Mohlke KL, Eriksson JG, Widén EE, Hattersley AT, Spector TD, Kähönen M, Viikari JS, Lehtimäki T, Boomsma DI, Sebert S, Vollenweider P, Sørensen TIA, Bisgaard H, Bønnelykke K, Murray JC, Melbye M, Nohr EA, Mook-Kanamori DO, Rivadeneira F, Hofman A, Felix JF, Jaddoe VWV, Hansen T, Pisinger C, Vaag AA, Pedersen O, Uitterlinden AG, Järvelin MR, Power C, Hyppönen E, Scholtens DM, Lowe WL Jr, Smith GD, Timpson NJ, Morris AP, Wareham NJ, Hakonarson H, Grant SFA, Frayling TM, Lawlor DA, Njølstad PR, Johansson S, Ong KK, McCarthy MI, Perry JRB, Evans DM, Freathy RM (2019) Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat Genet 51:804–814
Warrington NM, Freathy RM, Neale MC, Evans DM (2018) Using structural equation modelling to jointly estimate maternal and fetal effects on birthweight in the UK Biobank. Int J Epidemiol 47:1229–1241
Warrington NM, Richmond R, Fenstra B, Myhre R, Gaillard R, Paternoster L, Wang CA, Beaumont RN, Das S, Murcia M, Barton SJ, Espinosa A, Thiering E, Atalay M, Pitkänen N, Ntalla I, Jonsson AE, Freathy R, Karhunen V, Tiesler CMT, Allard C, Crawford A, Ring SM, Melbye M, Magnus P, Rivadeneira F, Skotte L, Hansen T, Marsh J, Guxens M, Holloway JW, Grallert H, Jaddoe VWV, Lowe WL Jr, Roumeliotaki T, Hattersley AT, Lindi V, Pahkala K, Panoutsopoulou K, Standl M, Flexeder C, Bouchard L, Aagaard Nohr E, Marina LS, Kogevinas M, Niinikoski H, Dedoussis G, Heinrich J, Reynolds RM, Lakka T, Zeggini E, Raitakari OT, Chatzi L, Inskip HM, Bustamante M, Hivert MF, Jarvelin MR, Sørensen TIA, Pennell C, Felix JF, Jacobsson B, Geller F, Evans DM, Lawlor DA (2018) Maternal and fetal genetic contribution to gestational weight gain. Int J Obes (Lond) 42:775–784
Wolf JB, Wade MJ (2009) What are maternal effects (and what are they not)? Philos Trans R Soc Lond B Biol Sci 364:1107–1115
Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM (2010) Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42:565–569
Yang J, Lee SH, Goddard ME, Visscher PM (2011a) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88:76–82
Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de Andrade M, Feenstra B, Feingold E, Hayes MG, Hill WG, Landi MT, Alonso A, Lettre G, Lin P, Ling H, Lowe W, Mathias RA, Melbye M, Pugh E, Cornelis MC, Weir BS, Goddard ME, Visscher PM (2011b) Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet 43:519–525
Yang J, Zaitlen NA, Goddard ME, Visscher PM, Price AL (2014) Advantages and pitfalls in the application of mixed-model association methods. Nat Genet 46:100–106
York TP, Eaves LJ, Lichtenstein P, Neale MC, Svensson A, Latendresse S, Långström N, Strauss 3rd JF (2013) Fetal and maternal genes’ influence on gestational age in a quantitative genetic analysis of 244,000 Swedish births. Am J Epidemiol 178:543–550
Young AI, Frigge ML, Gudbjartsson DF, Thorleifsson G, Bjornsdottir G, Sulem P, Masson G, Thorsteinsdottir U, Stefansson K, Kong A (2018) Relatedness disequilibrium regression estimates heritability without environmental bias. Nat Genet 50:1304–1310
Acknowledgements
We are extremely grateful to all the families who took part in ALSPAC, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. A comprehensive list of grants funding is available on the ALSPAC website (https://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf). The UK Medical Research Council and the Wellcome Trust (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. The ALSPAC GWAS data were generated by Sample Logistics and Genotyping Facilities at the Wellcome Trust Sanger Institute and LabCorp (Laboratory Corporation of America) using support from 23andMe. This publication is the work of the authors and DME will serve as guarantor for the contents of this paper.
Funding
NMW is supported by a National Health and Medical Research Council Early Career Fellowship (Grant No. ECF1104818). This work and DME are supported by an NHMRC Senior Research Fellowship (Grant No. SRF1137714) and NHMRC project grants (Grant Nos. GNT1085159, GNT1085130, GNT1125141, GNT1125200, GNT1157714). This work was supported by grants (to PRN) from the European Research Council (Grant No. AdG #293574), the Bergen Research Foundation (“Utilizing the Mother and Child Cohort and the Medical Birth Registry for Better Health”), Stiftelsen Kristian Gerhard Jebsen (Translational Medical Center), the University of Bergen, the Research Council of Norway (FRIPRO Grant #240413), the Western Norway Regional Health Authority (Strategic Fund “Personalized Medicine for Children and Adults”), and the Norwegian Diabetes Foundation; and (to SJ) Helse Vest's Open Research Grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Zhen Qiao, Jie Zheng, Øyvind Helgeland, Marc Vaudel, Stefan Johansson, Pål Njølstad, George Davey Smith, Nicole Warrington and David Evans declare that they have no conflict of interest.
Ethical approval
Ethical approval was obtained from the ALSPAC Law and Ethics Committee, MoBa Ethics Board and other relevant ethics committees. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Written informed consent has been provided by all study participants.
Additional information
Edited by Stacey Cherny.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Qiao, Z., Zheng, J., Helgeland, Ø. et al. Introducing M-GCTA a Software Package to Estimate Maternal (or Paternal) Genetic Effects on Offspring Phenotypes. Behav Genet 50, 51–66 (2020). https://doi.org/10.1007/s10519-019-09969-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10519-019-09969-4