Abstract
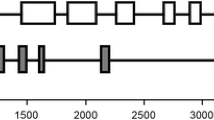
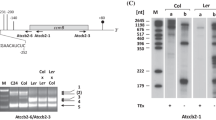
Partial amino acid sequences of a 49 kDa apyrase (ATP diphosphohydrolase, EC 3.6.1.5) from the cytoskeletal fraction of etiolated pea stems were used to derive oligonucleotide DNA primers to generate a cDNA fragment of pea apyrase mRNA by RT-PCR and these primers were used to screen a pea stem cDNA library. Two almost identical cDNAs differing in just 6 nucleotides within the coding regions were found, and these cDNA sequences were used to clone genomic fragments by PCR. Two nearly identical gene fragments containing 8 exons and 7 introns were obtained. One of them (H-type) encoded the mRNA sequence described by Hsieh et al. (1996) (DDBJ/EMBL/GenBank Z32743), while the other (S-type) differed by the same 6 nucleotides as the mRNAs, suggesting that these genes may be alleles. The six nucleotide differences between these two alleles were found solely in the first exon, and these mutation sites had two types of consensus sequences. These mRNAs were found with varying lengths of 3′ untranslated regions (3′-UTR). There are some similarities between the 3′-UTR of these mRNAs and those of actin and actin binding proteins in plants. The putative roles of the 3′-UTR and alternative polyadenylation sites are discussed in relation to their possible role in targeting the mRNAs to different subcellular compartments.
Similar content being viewed by others
Abbreviations
- cDNA:
-
complementary DNA
- CSB:
-
cytoskeleton-stabilizing buffer
- DTT:
-
dithiothreitol
- EDTA:
-
ethylenediaminetetraacetic acid
- HEPES:
-
N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid
- PMSF:
-
phenylmethylsulfonyl fluoride
- RT-PCR:
-
reverse transcription — polymerase chain reaction
- SDS:
-
sodium dodecyl sulfate
- Tris:
-
tris(hydroxymethyl)aminomethane
References
Etzler M.E., Kalsi G., Ewing N.N., Roberts N.J., Day R.B., Murphy J.B. 1999. A nod factor binding lectin with apyrase activity from legume roots. Proc. Natl. Acad. Sci. U.S.A., 96: 5856–5861.
Ghosh R., Biswas S., Roy S. 1998. An apyrase from Mimosa pudica contains N5,N10-methenyl tetrahydrofolate and is stimulated by light. Eur. J. Biochem., 258: 1009–1013
Handa M., Guidotti G. 1996. Purification and cloning of a soluble ATP-diphosphohydrolase (apyrase) from potato tubers (Solanum tuberosum). Biochem. Biophys. Res. Commun., 218: 916–923.
Hazelrigg T. 1998. The destinies and destinations of RNAs. Cell, 95: 451–460.
Hsieh H.L., Tong C.G., Thomas C., Roux S.J. 1996. Light-modulated abundance of an mRNA encoding a calmodulin-regulated, chromatin-associated NTPase in pea. Plant Mol. Biol., 30: 135–147.
Jansen R.-P. 1999. RNA-cytoskeletal associations. FASEB J., 13: 455–466.
Kislauskis E.H., Li Z., Singer R.H., Taneja K.L. 1993. Isoform-specific 3′-untranslated sequences sort α-cardiac and β-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J. Cell Biol., 123: 165–172
Kislauskis E.H., Zhu X., Singer R.H. 1994. Sequences responsible for intracellular localization of β-actin messenger RNA also affect cell phenotype. J. Cell Biol., 127: 441–451
Komoszyński M., Wojtczak A. 1996. Apyrases (ATP diphosphohydrolases, EC 3.6.1.5): function and relationship to ATPases. Biochim. Biophys. Acta., 1310: 233–241.
Matsumoto H., Yamaya T., Tanigawa M. 1984. Activation of ATPase activity in the calmodulin fraction of pea nuclei by calcium and calmodulin. Plant Cell Physiol., 25: 191–195
Meyerhof O. 1945. The origin of the reaction of Harden and Young in cell-free alcoholic fermentation. J. Biol. Chem., 157: 105–119.
Okita T.W., Li X, Roberts M.W. 1994. Targeting of mRNAs to domains of the endoplasmic reticulum. Trends Cell Biol., 4: 91–96.
O’Neil K.T., DeGrado W.F. 1990. How calmodulin binds its targets: sequence independent recognition of amphiphilic α-helicals. Trends Biochem. Sci., 15: 59–64.
Pabo C.O., Sauer R.T. 1992. Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem., 61: 1053–1095.
Pesole G., Liuni S., Grillo G., Ippedico M., Larizza A., Makalowski W., Saccone C. 1999. UTRdb: a specialized database of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Nucleic Acids Res., 27: 188–191.
Pinna L.A. 1990. Casein kinase 2: an ‘eminence grise’ in cellular regulation?. Biochem. Biophys. Acta, 1054: 267–284.
Raikhel N. 1990. Nuclear targeting in plants. Plant Physiol., 100: 267–284.
Roberts N.J., Brigham J., Wu B., Murphy J.B., Volpin H., Phillips D.A., Etzler M.E. 1999. A Nod factor-binding lectin is a member of a distinct class of apyrases that may be unique to the legumes. Mol. Gen. Genet., 262: 261–267.
Schroder H.C., Rottmann M., Bachmann M., Muller W.E. 1986. Purification and characterization of the major nucleoside triphosphate from rat liver nuclear envelopes. J. Biol. Chem., 261: 663–668.
Shibata K., Morita Y., Abe S. Stanković B., Davies E. 1999. Apyrase from pea stems: Isolation, purification, characterization and identification of a NTPase from the cytoskeleton fraction of pea stem tissue. Plant Physiol. Biochem., 37: 881–888.
Sundell C. L., Singer R.H. 1991. Requirement of microfilaments in sorting of actin messenger RNA. Science, 253: 1275–1277.
Thomas C., Sun Y., Naus K., Lloyd A., Roux S.J. 1999. Apyrase functions in plant phosphate nutrition and mobilizes phosphate from extracellular ATP. Plant Physiol., 119: 543–551.
Tognoli L, Marrè E. 1981. Purification and characterization of a divalent cation-activated ATPase-ADPase from pea stem microsomes. Biochim. Biophys. Acta. 642: 1–14.
Tong C.G., Dauwalder M., Clawson G.A., Hatem C.L., Roux S.J. 1993. The major nucleoside triphosphatase in pea (Pisum sativum L.) nuclei and in rat liver nuclei share common epitopes also present in nuclear lamins. Plant Physiol., 101: 1005–1010.
Vara K., Serrano R. 1981. Purification and characterization of a membrane-bound ATP diphosphohydrolase from Cicer arieteum (chick pea) roots. Biochem. J., 197: 637–643
Wang T.F., Guidotti G. 1996. CD39 is an ecto-(Ca2+,Mg2+)-apyrase. J. Biol. Chem., 271:9898–9901.
Author information
Authors and Affiliations
Additional information
Sequence data from this article were deposited with the DDBJ/EMBL/GenBank Data Libraries under Accession Nos. Genomic sequences of pea apyrase: AB023621, AB030444, AB030445, AB038554, AB038555. cDNA sequences of pea apyrase: AB022319, AB027614, AB038668, AB038669.
Rights and permissions
About this article
Cite this article
Shibata, K., Abe, S. & Davies, E. Structure of the coding region and mRNA variants of the apyrase gene from pea (Pisum sativum). Acta Physiol Plant 23, 3–13 (2001). https://doi.org/10.1007/s11738-001-0016-y
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s11738-001-0016-y