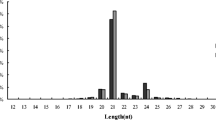
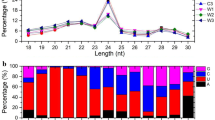
Abstract
miRNAs (microRNAs) are ~ 21-nt non-coding small RNAs (sRNAs) that play crucial regulatory roles in plant biotic and abiotic stress responses. Phosphorus (Pi) deficiency constrains plant growth and reduces yields worldwide. To identify tree miRNAs and evaluate their functions in the response to low Pi, we identified 261 known and 31 candidate novel miRNA families from three sRNA libraries constructed from Populus tomentosa subjected to sufficient or Pi deficiency condition or to restoration of a sufficient Pi level after Pi deficiency. Pi deficiency resulted in significant changes in the abundance of TPM (transcript per million) of 65 known and 3 novel miRNAs. Interestingly, four miRNAs responsive to low N—miR167, miR394, miR171, and miR857—were found to be involved in the response to low Pi. Thirty-five known and one novel miRNAs responded dynamically to Pi fluctuations, suggesting their involvement in the response to Pi deficiency. miRNA clusters comprising 36 miRNAs were identified in 10 chromosomes. Intriguingly, nine pairs of sense and antisense miRNAs transcribed from the same loci were detected in P. tomentosa, which is the first such report in woody plants. Moreover, target genes of the known miRNAs and novel miRNA candidates with significantly changed abundance were predicted, and their functions were annotated. Degradome sequencing supported the identified targets of miRNAs in P. tomentosa. These findings will enhance our understanding of universal and specific molecular regulatory mechanisms of trees under nutrition stress and may facilitate improvement of the Pi utilization efficiency of woody plants.






Similar content being viewed by others
References
Abdel-Ghany SE, Pilon M (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J Biol Chem 283:15932–15945
Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18:758–762
Akpinar BA, Kantar M, Budak H (2015) Root precursors of microRNAs in wild emmer and modern wheats show major differences in response to drought stress. Funct Integr Genomics 15:587–598
Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207–221
Alptekin B, Akpinar BA, Budak H (2017) A comprehensive prescription for plant miRNA identification. Front Plant Sci 7:2058
Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H (2005) Clustering and conservation patterns of human microRNAs. Nucleic Acids Res 33:2697–2706
Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141:1000–1011
Bari R, Datt PB, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141:988–999
Barik S, Kumar A, Sarkar DS, Yadav S, Gautam V, Singh A, Singh S, Sarkar AK (2015) Coevolution pattern and functional conservation or divergence of miR167s and their targets across diverse plant species. Sci Rep 5:14611
Budak H, Akpinar BA (2015) Plant miRNAs: biogenesis, organization and origins. Funct Integr Genomics 15:523–531
Chen L, Ren Y, Zhang Y, Xu J, Sun F, Zhang Z, Wang Y (2012a) Genome-wide identification and expression analysis of heat-responsive and novel microRNAs in Populus tomentosa. Gene 504:160–165
Chen L, Ren Y, Zhang Y, Xu J, Zhang Z, Wang Y (2012b) Genome-wide profiling of novel and conserved Populus microRNAs involved in pathogen stress response by deep sequencing. Planta 235:873–883
Chen L, Zhang Y, Ren Y, Xu J, Zhang Z, Wang Y (2012c) Genome-wide identification of cold-responsive and new microRNAs in Populus tomentosa by high-throughput sequencing. Biochem Biophys Res Commun 417:892–896
Chiou TJ (2007) The role of microRNAs in sensing nutrient stress. Plant Cell Environ 30:323–332
Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL (2006) Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell 18:412–421
Cui X, Xu SM, Mu DS, Yang ZM (2009) Genomic analysis of rice microRNA promoters and clusters. Gene 431:61–66
Duan Y, Li W, Wu W, Pan R, Zhou Y, Qi J, Lin L, Chen Z, Mao D, Liu H, Zhang D, Xue Y (2003) Genetic analysis and mapping of gene fzp(t) controlling spikelet differentiation in rice. Sci China C Life Sci 46:328–334
Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15:2038–2043
German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, De Paoli E, Lu C, Schroth G, Meyers BC, Green PJ (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26:941–946
Greb T, Clarenz O, Schafer E, Muller D, Herrero R, Schmitz G, Theres K (2003) Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17:1175–1187
Hackenberg M, Shi BJ, Gustafson P, Langridge P (2013) Characterization of phosphorus-regulated miR399 and miR827 and their isomirs in barley under phosphorus-sufficient and phosphorus-deficient conditions. BMC Plant Biol 13:214
Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151:2120–2132
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14:787–799
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484
Kant S, Peng M, Rothstein SJ (2011) Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet 7:e1002021
Kantar M, Lucas SJ, Budak H (2011) miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 233:471–484
Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi H, Dalmay T (2009) Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J 57:313–321
Kurtoglu KY, Kantar M, Lucas SJ, Budak H (2013) Unique and conserved MicroRNAs in wheat chromosome 5D revealed by next-generation sequencing. PLoS One 8:e69801
Laloi C, Apel K, Danon A (2004) Reactive oxygen signalling: the latest news. Curr Opin Plant Biol 7:323–328
Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks DS, Sander C, Tuschl T, Gaul U (2005) Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell 121:1097–1108
Li R, Li Y, Kristiansen K, Wang J (2008a) SOAP: short oligonucleotide alignment program. Bioinformatics 24:713–714
Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK (2008b) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20:2238–2251
Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R (2010) Transcriptome-wide identification of microRNA targets in rice. Plant J 62:742–759
Li B, Qin Y, Duan H, Yin W, Xia X (2011) Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J Exp Bot 62:3765–3779
Liang G, He H, Yu D (2012) Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS One 7:e48951
Lin WY, Huang TK, Chiou TJ (2013) Nitrogen limitation adaptation a target of MicroRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 25:4061–4074
Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ (2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24:2168–2183
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408
Lu S, Sun YH, Shi R, Clark C, Li L, Chiang VL (2005) Novel and mechanical stress-responsive MicroRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17:2186–2203
Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, Griffiths-Jones S, Jacobsen SE, Mallory AC, Martienssen RA, Poethig RS, Qi Y, Vaucheret H, Voinnet O, Watanabe Y, Weigel D, Zhu JK (2008) Criteria for annotation of plant MicroRNAs. Plant Cell 20:3186–3190
Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, Eaves CJ, Marra MA (2008) Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18:610–621
Neill SJ, Desikan R, Clarke A, Hurst RD, Hancock JT (2002) Hydrogen peroxide and nitric oxide as signaling molecules in plants. J Exp Bot 53:1237–1247
Nguyen GN, Rothstein SJ, Spangenberg G, Kant S (2015) Role of microRNAs involved in plant response to nitrogen and phosphorous limiting conditions. Front Plant Sci 6:629
Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR (2009) Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol 150:1541–1555
Park BS, Seo JS, Chua NH (2014) NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26:454–464
Pei H, Ma N, Chen J, Zheng Y, Tian J, Li J, Zhang S, Fei Z, Gao J (2013) Integrative analysis of miRNA and mRNA profiles in response to ethylene in rose petals during flower opening. PLoS One 8:e64290
Ren Y, Chen L, Zhang Y, Kang X, Zhang Z, Wang Y (2012) Identification of novel and conserved Populus tomentosa microRNA as components of a response to water stress. Funct Integr Genomics 12:327–339
Ren Y, Sun F, Hou J, Chen L, Zhang Y, Kang X, Wang Y (2015) Differential profiling analysis of miRNAs reveals a regulatory role in low N stress response of Populus. Funct Integr Genomics 15:93–105
Rufty TW, Mackown CT, Israel DW (1990) Phosphorus stress effects on assimilation of nitrate. Plant Physiol 94:328–333
Schwab R, Voinnet O (2009) miRNA processing turned upside down. EMBO J 28:3633–3634
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8:517–527
Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J (2004) A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res 14:1741–1748
Shin R, Berg RH, Schachtman DP (2005) Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol 46:1350–1357
Shuai P, Liang D, Zhang Z, Yin W, Xia X (2013) Identification of drought-responsive and novel Populus trichocarpa microRNAs by high-throughput sequencing and their targets using degradome analysis. BMC Genomics 14:233
Si J, Zhou T, Bo W, Xu F, Wu R (2014) Genome-wide analysis of salt-responsive and novel microRNAs in Populus euphratica by deep sequencing. BMC Genet 15(Suppl 1):S6
Song Y, Wang Z, Bo W, Ren Y, Zhang Z, Zhang D (2012) Transcriptional profiling by cDNA-AFLP analysis showed differential transcript abundance in response to water stress in Populus hopeiensis. BMC Genomics 13:286
Sun X, Xie Z, Zhang C, Mu Q, Wu W, Wang B, Fang J (2016) A characterization of grapevine of GRAS domain transcription factor gene family. Funct Integr Genomics 16:347–363
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Tanzer A, Stadler PF (2004) Molecular evolution of a microRNA cluster. J Mol Biol 339:327–335
Valdes-Lopez O, Arenas-Huertero C, Ramirez M, Girard L, Sanchez F, Vance CP, Luis RJ, Hernandez G (2008) Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ 31:1834–1843
Wang P, Grimm B (2015) Organization of chlorophyll biosynthesis and insertion of chlorophyll into the chlorophyll-binding proteins in chloroplasts. Photosynth Res 126:189–202
Wang C, Shangguan L, Kibet KN, Wang X, Han J, Song C, Fang J (2011) Characterization of microRNAs identified in a table grapevine cultivar with validation of computationally predicted grapevine miRNAs by miR-RACE. PLoS One 6:e21259
Wang Y, Zhang C, Hao Q, Sha A, Zhou R, Zhou X, Yuan L (2013) Elucidation of miRNAs-mediated responses to low nitrogen stress by deep sequencing of two soybean genotypes. PLoS One 8:e67423
Xu J, Wong C (2008) A computational screen for mouse signaling pathways targeted by microRNA clusters. RNA 14:1276–1283
Xu F, Liu Q, Chen L, Kuang J, Walk T, Wang J, Liao H (2013) Genome-wide identification of soybean microRNAs and their targets reveals their organ-specificity and responses to phosphate starvation. BMC Genomics 14:66
Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T (2009) SQUAMOSA promoter binding protein-Like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21:347–361
Yu J, Wang F, Yang GH, Wang FL, Ma YN, Du ZW, Zhang JW (2006) Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem Biophys Res Commun 349:59–68
Zeng HQ, Zhu YY, Huang SQ, Yang ZM (2010) Analysis of phosphorus-deficient responsive miRNAs and cis-elements from soybean (Glycine max L.). J Plant Physiol 167:1289–1297
Zhang B, Pan X, Stellwag EJ (2008) Identification of soybean microRNAs and their targets. Planta 229:161–182
Zhang Y, Zhang R, Su B (2009) Diversity and evolution of MicroRNA gene clusters. Sci China C Life Sci 52:261–266
Zhang D, Song H, Cheng H, Hao D, Wang H, Kan G, Jin H, Yu D (2014) The acid phosphatase-encoding gene GmACP1 contributes to soybean tolerance to low-phosphorus stress. PLoS Genet 10:e1004061
Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, Ruan K, Jin Y (2007) Identification of drought-induced microRNAs in rice. Biochem Biophys Res Commun 354:585–590
Zhao M, Tai H, Sun S, Zhang F, Xu Y, Li WX (2012) Cloning and characterization of maize miRNAs involved in responses to nitrogen deficiency. PLoS One 7:e29669
Zhou L, Chen J, Li Z, Li X, Hu X, Huang Y, Zhao X, Liang C, Wang Y, Sun L, Shi M, Xu X, Shen F, Chen M, Han Z, Peng Z, Zhai Q, Chen J, Zhang Z, Yang R, Ye J, Guan Z, Yang H, Gui Y, Wang J, Cai Z, Zhang X (2010) Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS One 5:e15224
Data archiving statement
The raw sequence data can be available at the NIH Short Read Archive database (http://www.ncbi.nlm.nih.gov/sra) under the BioProject ID PRJNA430018 (small RNA sequences) and PRJNA430019 (degradome sequences).
Funding
This work was supported by the Fundamental Research Funds for Central Universities (No.2015ZCQ-SW-01), National Natural Science Foundation of China (No. 31670671, 31470668).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Suppl. Fig. S1
Mature and precursor sequences and predicted stem-loop structures of novel miRNAs from Populous tomentosa. The sequences indicated in red color are miRNAs sequences, and sequences in green color are miRNA* sequences (PDF 3467 kb)
Suppl. Fig. S2
Target plots (t-plots) of miRNA targets in different categories confirmed by degradome sequencing. The normalized signature abundance throughout the length of the indicated transcripts is shown. Representative t-plots for class 0 (a), class 1 (b), class 2 (c), class 3 (d), and class 4 (e) categories are shown. Red peaks indicate signatures consistent with miRNA-directed cleavage. The solid lines and dot in miRNA:mRNA alignments indicate matched RNA base pairs and GU mismatch, respectively. On the left of the t-plots, the cleavage sites and raw tags are shown in the left and right column, respectively (PDF 210 kb)
Suppl. Fig. S3
GO enrichment analysis for the target genes of miRNAs with altered expression in P. tomentosa under Pi starvation stress. (a) GO enrichment of targets genes of miRNAs with altered expression in DP/KK. (b) GO enrichment of targets genes of miRNAs with altered expression in HF/DP (PDF 151 kb)
Table S1
(XLS 22 kb)
Table S2
(XLSX 9 kb)
Table S3
(DOCX 14 kb)
Table S4
(XLSX 23 kb)
Table S5
(XLS 131 kb)
Table S6
(XLSX 17 kb)
Table S7
(XLS 73 kb)
Table S8
(DOC 34 kb)
Table S9
(DOC 162 kb)
Table S10
(XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Bao, H., Chen, H., Chen, M. et al. Transcriptome-wide identification and characterization of microRNAs responsive to phosphate starvation in Populus tomentosa. Funct Integr Genomics 19, 953–972 (2019). https://doi.org/10.1007/s10142-019-00692-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-019-00692-1