Abstract
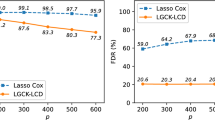
Motivated by high-dimensional genomic studies, we develop an improved procedure for adaptive Lasso in high-dimensional survival analysis. The proposed procedure effectively reduces the false discoveries while successfully maintaining the false negative proportions, which improves the existing adaptive Lasso procedures. The implementation of the proposed procedure is straightforward and it is sufficiently flexible to accommodate large-scale problems where traditional procedures are impractical. To quantify the uncertainty of variable selection and control the family-wise error rate, a multiple sample-splitting based testing algorithm is developed. The practical utility of the proposed procedure are examined through simulation studies. The methods developed are then applied to a multiple myeloma data set.


Similar content being viewed by others
References
Alexande DH, Lange K (2011) Stability selection for genome-wide association. Genet Epidemiol 35(7):722–728
Bataille R, Grenier J, Sany J (1984) Beta-2-microglobulin in myeloma: optimal use for staging, prognosis, and treatment-a prospective study of 160 patients. Blood 63(2):468–476
Bühlmann P, van de Geer S (2011) Statistics for high-dimensional data: methods, theory and applications. Springer, Berlin
Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Golub TR (2011) Initial genome sequencing and analysis of multiple myeloma. Nature 471(7339):467–472
Di Luccio E (2015) Inhibition of nuclear receptor binding SET domain 2/multiple myeloma SET domain by LEM-06 implication for epigenetic cancer therapies. J Cancer Prev 20(2):113–120
Fan J, Li R (2001) Variable selection via nonconcave penalized likelihood and its oracle properties. J Am Stat Assoc 96(456):1348–1360
Fan J, Li R (2002) Variable selection for Cox’s proportional hazards model and frailty model. Ann Stat 30(1):74–99
Geoman JJ (2010) L1 penalized estimation in the Cox proportional hazards model. Biom J 52(1):70–84
Gui J, Li H (2005) Penalized cox regression analysis in the high-dimensional and low-sample size settings with application to microarray gene expression data. Bioinformatics 21(13):3001–3008
Hastie T, Tibshirani R, Friedman J (2009) The elements of statistical learning: data mining, inference, and prediction. Springer, New York
Heagerty PJ, Zheng Y (2005) Survival model predictive accuracy and ROC curves. Biometrics 61(1):92105
Kyle RA, Rajkuma SV (2008) Multiple myeloma. Blood 111(6):2962–2972
MAQC Consortium (2010) The MAQC-II project: a comprehensive study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol 28(8):827–838
Meinshausen N, Meier L, Bühlmann P (2009) P-values for high-dimensional regression. J Am Stat Assoc 104(488):1671–1681
Shaughnessy JD, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, Stewart JP, Kordsmeier B, Randolph C, Williams DR, Xiao Y, Xu H, Epstein J, Anaissie E, Krishna SG, Cottler-Fox M, Hollmig K, Mohiuddin A, Pineda-Roman M, Tricot G, van Rhee F, Sawyer J, Alsayed Y, Walker R, Zangari M, Crowley J, Barlogie B (2007) A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood 109(6):2276–2284
Simon N, Friedman J, Hastie T, Tibshirani R (2011) Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw 39(5):1–13
Song LL, Ponomareva L, Shen H, Duan X, Alimirah F, Choubey D (2010) Interferon-inducible IFI16, a negative regulator of cell growth, down-regulates expression of human telomerase reverse transcriptase (hTERT) gene. PLOS ONE 5(1):e8569
Sun S, Hood M, Scott L, Peng Q, Mukherjee S, Tung J, Zhou X (2017) Differential expression analysis for RNAseq using Poisson mixed models. Nucleic Acids Res 45(11):e106
Tibshirani R (1996) Regression shrinkage and selection via the lasso. J R Stat Soc Ser B (Methodol) 58(1):267–288
Tibshirani R (1997) The lasso method for variable selection in the Cox model. Stat Med 16(4):385–395
Uno H, Cai T, Pencina MJ, D‘gostino RB, Wei LJ (2011) On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med 30(10):1105–1117
Zhang H, Lu W (2007) Adaptive Lasso for Cox’s proportional hazards model. Biometrika 94(3):691–703
Zhao DS, Li Y (2014) Score test variable screening. Biometrics 70(4):862–871
Zhou SH, van de Geer S, Bühlmann P (2009) Adaptive Lasso forhigh dimensional regression and Gaussian graphical modeling. arXiv:0903.2515
Zou H, Hastie T (2005) Regression shrinkage and selection via the elastic net with application to microarrays. J R Stat Soc Ser B (Methodol) 67(2):301–320
Zou H, Li R (2008) One-step sparse estimates in nonconcave penalized likelihood models. Ann Stat 36(4):1509–1533
Zou H, Zhang HH (2009) On the adaptive elastic-net with a diverging number of parameters. Ann Stat 37(4):1733–1751
Author information
Authors and Affiliations
Corresponding author
Appendix: FWER-control procedure in Sect. 3.3
Appendix: FWER-control procedure in Sect. 3.3
-
(a)
Randomly split the original data multiple times (say B). Specifically, for \(b=1, \ldots , B\), split the data into two disjoint sets with sample size \(n_1=\lfloor n/2 \rfloor \) and \(n_2=n-\lfloor n/2 \rfloor \), respectively. Here \(\lfloor n/2 \rfloor \) is defined as the largest integer not greater than n / 2.
-
(b)
For \(b=1, \ldots , B\), select variables based on the first half of the data and denote the index set of selected variables by \({\widehat{{{\mathcal {S}}}}}^{(b)}.\)
-
(c)
Based on the second half of the data, fit conventional Cox model and assign p-values. denoted by \({\tilde{P}}_{j}\) for \(j=1,\dots ,p\), using variables selected from step (b). For variables not selected from the first half of the data, assign their p-values as 1.
-
(d)
Compute adjusted p-values to correct for the multiplicity of the testing problem
$$\begin{aligned} {\tilde{P}}_{corrected,j} = min({\tilde{P}}_{j} |{\widehat{{{\mathcal {S}}}}}^{(b)}|,1), \end{aligned}$$where \(|\widehat{{\mathcal {S}}}^{(b)}|\) is the cardinality, e.g., number of variables in \(\widehat{{\mathcal {S}}}^{(b)}\).
-
(e)
To aggregate the adjusted p-values over multiple splitting (e.g., B values for each covariate), define
$$\begin{aligned} Q_{j}(\gamma ) = min \{q_{\gamma }( \{ {\tilde{P}}^{[b]}_{corrected,j}/\gamma ; b=1,\ldots ,B \}),1 \} \end{aligned}$$where \(\gamma \in (0,1)\) and \(q_{\gamma }\) is the emperical \(\gamma \)-quantile function. Define the final p-values as
$$\begin{aligned} P_{j} = min \{(1-log \gamma _{min}) \underset{\gamma \in (\gamma _{min},1)}{\inf } Q_{j}(\gamma ),1 \}, \end{aligned}$$where \(\gamma _{min} \in (0,1)\) is a lower bound for \(\gamma \), typically 0.05 (Meinshausen et al. 2009).
Rights and permissions
About this article
Cite this article
He, K., Wang, Y., Zhou, X. et al. An improved variable selection procedure for adaptive Lasso in high-dimensional survival analysis. Lifetime Data Anal 25, 569–585 (2019). https://doi.org/10.1007/s10985-018-9455-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10985-018-9455-2