Abstract
Background
Internode elongation is an important agronomic trait in rice that determines culm length, which is related to lodging, panicle exsertion, and biomass. sui4 (shortened uppermost internode 4) mutants show reduced internode length and a dwarf phenotype due to shortened internodes; the uppermost internode is particularly severely affected. The present study was performed to identify the molecular nature and function of the SUI4 gene during internode elongation.
Results
Our previous study showed that the SUI4 gene was mapped to a 1.1-Mb interval on chromosome 7 (Ji et al. 2014). In order to isolate the gene responsible for the sui4 phenotype, genomic DNA resequencing of sui4 mutants and wild-type plants and reciprocal transformation of wild-type and mutant alleles of the putative SUI4 gene was performed. The data revealed that the causative mutation of sui4 was a T to A nucleotide substitution at the microRNA172 binding site of Os07g0235800, and that SUI4 is a new allele of the previously reported gene SUPERNUMERARY BRACT (SNB), which affects flower structure. In order to understand the effect of this mutation on expression of the SUI4/SNB gene, SUI4/SNB native promoter-fuzed GUS transgenics were examined, along with qRT-PCR analysis at various developmental stages. In sui4 mutants, the SUI4/SNB gene was upregulated in the leaves, culms, and panicles, especially when internodes were elongated. In culms, SUI4/SNB was expressed in the nodes and the lower parts of elongating internodes. In order to further explore the molecular nature of SUI4/SNB during internode elongation, RNA-seq and qRT-PCR analysis were performed with RNAs from the culms of sui4 mutants and wild-type plants in the booting stage. The data showed that in sui4 mutants, genes deactivating bioactive gibberellins and cytokinin were upregulated while genes related to cell expansion and cell wall synthesis were downregulated.
Conclusion
In summary, this paper shows that interaction between SUI4/SNB and microRNA172 could determine internode elongation during the reproductive stage in rice plants. Due to a mutation at the microRNA172 binding site in sui4 mutants, the expression of SUI4/SNB was enhanced, which lowered the activities of cell expansion and cell wall synthesis and consequently resulted in shortened internodes.
Similar content being viewed by others
Background
The culm of rice plants is composed of solid nodes and hollow internodes. Internode elongation is an important agronomic trait that determines culm length, which is related to lodging, panicle exsertion, and biomass. Rice internodes can be divided into three regions: the intercalary meristem (IM), the elongation zone (EZ), and the differentiation zone (DZ) (Kende et al. 1998). At the base of the internode is the IM, where cell division generates new internodal cells. These new cells are displaced into the EZ, where they elongate until they reach their final length, and growth ceases in the DZ, where formation of the secondary wall and xylem takes place (Kende et al. 1998).
Gibberellin (GA) activates the cell cycle in the IM as well as cell elongation (Kende et al. 1998). Mutations in genes involved in GA biosynthesis and signaling hamper internode elongation and cause dwarfism, as exemplified in rice sd1, d18, d35, d1, gid1, and gid2 mutants. GA signals promote cellulose synthesis by preventing interaction between SLENDER RICE1 (SLR1), a DELLA repressor of GA signaling, and NACs, the top-layer transcription factors for secondary wall formation (Huang et al. 2015).
Brassinosteroids (BRs) are also essential in stem elongation (Je et al. 2010). Under physiological conditions, BRs promote GA accumulation by regulating the expression of GA metabolic genes to stimulate cell elongation (Tong et al. 2014). BRs increase the expression of two bHLH transcription factors, ILI1 (INCREASED LAMINA INCLINATION 1) and PRE1 (PACLOBUTRAZOL RESISTANCE 1), which both increase cell elongation, but repress the bHLH transcription factor IBH1 (ILI1 BINDING BHLH 1) through BZR1 (BRASSINAZOLE-RESISTANT 1), which suppresses cell elongation (Zhang et al. 2009). Several rice dwarf mutants had mutations in genes involved in BR biosynthesis and signaling; they are d2, d11, brd1, brd2, d61, and dlt.
The main culm of japonica rice has 14–17 nodes; internode elongation occurs only above the fifth node from the top, while the lower internodes remain unelongated (Chonan 1993). The uppermost internode is the longest, and lower internodes become progressively shorter (Chonan 1993). Upon shifting from vegetative to reproductive growth, four or five upper internodes (UPIs) sequentially elongate from the basal end to the uppermost internode. This process is concomitant with panicle development, suggesting that signaling occurs between panicles and UPIs during the reproductive phase (Itoh et al. 2005; Wang et al. 2009; Yamamuro et al. 2000). Identification of regulators of UPI elongation and floral development should provide clues as to the underlying genetic factors that coordinate organogenesis in both vegetative phytomers and panicles (Wang et al. 2009).
microRNAs play important roles in controlling plant development, productivity, and defense against biotic and abiotic stresses by negatively regulating gene expression at the post-transcriptional level (Teotia and Tang 2015). miR172 regulates the expression of a small group of AP2-like transcription factors and the transitions between developmental stages, as well as functioning in the specification of floral organ identity (Zhu and Helliwell 2011). Rice miR172 induces flowering by suppressing OsIDS1 (Oryza sativa INDETERMINATE SPIKELET1) and SNB (SUPERNUMERARY BRACT), two AP2 genes that negatively regulate the expression of Ehd1 (EARLY HEADING DATE 1) (Lee et al. 2014). The SNB gene, which is one of targets of miR172, controls the transition from spikelet meristem to floral meristem; in SNB knockout plants, the transition from spikelet meristems to floral meristems is delayed, resulting in the production of multiple rudimentary glumes (Lee et al. 2007). Overexpression of miR172b in rice has been shown to delay the transition from spikelet meristem to floral meristem, resulting in floral and seed developmental defects including changes in the number and identity of floral organs (Zhu et al. 2009).
In our previous study, we characterized sui4 mutant rice plants (Ji et al. 2014). The uppermost internodes of the sui4 mutants were severely shortened, which caused enclosure of panicles in the flag leaf sheath. The cells in these uppermost internodes were also remarkably shorter and more densely packed than those in the wild type (Dongjin). The SUI4 gene showed incomplete dominance or semidominance and was mapped to a 1.1-Mb region, RM1253-S0701579, on chromosome 7.
In the present study, we identified the molecular nature of the sui4 mutation by resequencing of mutant and wild-type genomic DNA and by reciprocal transformation of SUI4 alleles. It was revealed that a single nucleotide substitution in the miR172 binding site resulted in the dwarf phenotype and that SUI4 was allelic to SNB. To understand the effect of the mutation on expression of the SUI4/SNB gene, SUI4/SNB native promoter-fuzed GUS transgenics and qRT-PCR were employed. RNA-seq analysis demonstrated that SUI4/SNB could affect the homeostasis of GA and cytokinin. This study is the first to show that the expression interaction between the AP2 domain gene and miR172 determines the elongation of upper internodes in rice.
Results
A point mutation in SUI4 causes upregulation of this gene by blocking miRNA172 binding
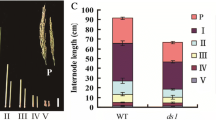
Relative to wild-type (Dongjin) plants, sui4 mutants showed reduced internode length and eventually a dwarf phenotype. In the mutants, the uppermost internode was much more severely shortened than the lower internodes (Fig.1a, b, c). The length of individual cells in the uppermost internode was also much shorter in sui4 mutants than in Dongjin plants (Fig. 1d, e).
Phenotype comparison between Dongjin plants and sui4 mutants. (a) Plant architecture at maturing stage. (b) Culm structure. Arrows indicate locations of nodes. Two main culms from two plants are shown for each phenotype. (c) Comparison of internode length between Dongjin plants and sui4 mutants. PL: panicle length, I1L: first internode length, I2L: second internode length, I3L: third internode length, I4L: fourth internode length, CL: culm length. (d) Longitudinal sections of the uppermost internodes of Dongjin plants and sui4 mutants. Red bars denote 50 μm. HP: hollow pith, OS: outer space, Sc: sclerenchyma tissue, Pc: parenchyma tissue. (e) Cell length in the uppermost internodes of Dongjin plants and sui4 mutants. The lengths of cells in parenchyma tissues were measured. Data are shown as the mean ± s.d. (t-test, *** P < 0.001). The cells in three samples were measured for Dongjin plants and sui4 mutants, respectively
To identify the locus responsible for the sui4 phenotype, sui4 mutants and Dongjin plants were subjected to genomic resequencing. Twenty-two SNPs were identified in the 1.1-Mb interval region mapped in our previous study (Additional file 1: Table S1, Fig. 2) (Ji et al. 2014). Only six SNPs were located in genic regions, while the rest were in intergenic regions. Among the six genic SNPs, five were located within introns, while only one was in the coding sequence of the locus Os07g0235800. The SNP in the coding region created a missense mutation that resulted in a single amino acid substitution from serine to threonine. The expression of five genes harboring all six genic SNPs was examined by RT-PCR. Except for Os07g0235800, genes carrying SNPs within introns were expressed at the same levels in Dongjin plants and sui4 mutants (Additional file 4: Figure S1). Since the five SNPs within introns did not seem to affect expression levels, it is likely that they are not responsible for the mutation of SUI4. Thus, Os07g0235800, which was expressed at higher levels in sui4 mutants than in Dongjin plants, was suspected to be the most probable candidate gene for the SUI4 phenotype.
SNPs found in the SUI4 gene region on chromosome 7. Twenty-two SNPs were identified between sui4 and Dongjin in the 1.1- Mb interval region (RM1253-S07015) mapped in our previous study (Ji et al. 2014). Detailed information was shown in Additional file 1: Table S1. Red arrows indicate the positions of SNPs in genic regions while blue arrows indicate the positions of SNPs in intergenic regions
Interestingly, the SNP was located at the miR172 binding site of Os07g0235800, leading to the hypothesis that the SNP might disrupt the affinity of miR172 to the binding site (Fig. 3a), which would weaken miR172-mediated suppression of Os07g0235800. To explore the effect of the putative miR172 binding site on the expression of Os07g0235800, the expression patterns of Os07g0235800 were compared at different developmental stages (seedling, booting, and heading stages) and in different tissues (leaves, roots, and culms) between Dongjin plants and sui4 mutants (Fig. 3b). Only in the leaves of two-week-old seedlings, booting stage panicles,and heading stage culms were the expression levels of SUI4 similar between Dongjin plants and sui4 mutants. In all other samples, levels of SUI4 mRNA were higher in sui4 mutants than in Dongjin plants. Expression was particularly high in sui4 mutant culms in the booting stage, when the culm was growing rapidly. Meanwhile, during the heading stage, when culm growth ceased, SUI4 expression was higher in sui4 mutant flag leaves and panicles than in culms. In Dongjin plants, the highest level of SUI4 mRNA expression was in the leaves of 70-day-old plants. This comparative expression study indicates that the expression of Os07g0235800 in Dongjin plants might be affected by the miR172 binding site, which, in sui4 mutants, harbored a point mutation. Overall, the difference in Os07g0235800 expression between Dongjin plants and sui4 mutants became more pronounced in the later stages of development, when the culm was growing rapidly. This is in agreement with previous reports of microRNA172 expression increasing as plants grow (Zhu and Helliwell 2011; Zhu et al. 2009).
Os07g0235800 gene mutation and expression pattern. (a) Structure of the Os07g0235800 gene and location of mutation. Empty boxes indicate 5′ and 3′ UTRs, filled boxes indicate CDS, and lines indicate introns. (b) Expression pattern of Os07g0235800 at different developmental stages and organs in Dongjin plants and sui4 mutants. Relative fold expression difference is based on the expression level detected in two-week-old leaves as a baseline. Error bars represent standard deviation of the expression ratio. 2WL: two-week-old seedling leaf. 2WR: two-week-old seedling root. 4WL: four-week-old seedling leaf. ML: middle stage (70-day-old plant) leaf, BTFL: flag leaf of booting stage plant, BTPA: panicle of booting stage plant, BTCL: culm of booting stage plant, HDFL: flag leaf of heading stage plant, HDPA: panicle of heading stage plant, HDCL: culm of heading stage plant
In order to clarify the genetic relationship between the wild-type SUI4 allele of Dongjin plants and the SUI4 allele of the sui4 mutant, genomic DNA containing Os07g0235800 was isolated from Dongjin plants and sui4 mutants and was reciprocally transformed into sui4 mutants and Dongjin plants, respectively. First, a vector harboring the 7.9-kb genomic DNA that contained the genic region of wild-type Os07g0235800, including its promoter and terminator, was transformed from a Dongjin plant into sui4 mutants. The resulting transgenic line, referred to as sui4-DJ, was not significantly different from sui4 mutants (Fig. 4a-d, Additional file 5: Figure S2). This may be due to the fact that SUI4 shows semi-dominant characteristics (Ji et al. 2014). In contrast, transformation of a vector harboring a 7.9-kb genic region of Os07g0235800, including its promoter and terminator, from a sui4 mutant into Dongjin plants caused a remarkable reduction in internode length in the resulting transgenic line, DJ-SUI4 (Fig. 4a–d, Additional file 5: Figure S2). This demonstrates that Os07g0235800 is responsible for the sui4 phenotype.
Mutations in the Os07g0235800 gene and phenotypes of transgenic plants. (a) Structure of the SUI4/SNB gene and location of mutations. Empty boxes indicate 5′ and 3′ UTRs, filled boxes indicate CDS, and lines indicates introns. (b) Plant architecture at the maturing stage. DJ: Dongjin, sui4-DJ: transgenic line in which the SUI4 gene from a Dongjin plant was introduced into sui4 mutants, DJ-SUI4: transgenic line in which the SUI4 gene from a sui4 mutant was introduced into Dongjin plants, DJ-SUI4sd: transgenic line in which the SUI4 gene, modified by site-directed mutagenesis as shown in (a), was introduced into Dongjin plants. (c) Culm structure. Arrows indicate locations of nodes. Two main culms from two plants are shown for each phenotype. Two independent transgenic lines are shown for DJ-SUI4 and DJ-SUI4sd. (d) Lengths of panicles, culms, and internodes. For DJ and sui4, three main culms from three different plants were measured. For sui4-DJ, DJ-SUI4, and DJ-SUI4sd, six main culms from three to four independent transgenic lines were measured. PL: panicle length, I1L: first internode length, I2L: second internode length, I3L: third internode length, I4L: forth internode length, CL: culm length
Next, since sui4 mutants harbored the single T to A nucleotide substitution, which resulted in one amino acid substitution from serine (TCA) to threonine (ACA), an experiment was performed to determine whether the mutant phenotype arose due to a single amino acid substitution or due to the single nucleotide change in the miR172 binding site. A vector was constructed in which the amino acid threonine (ACA) of sui4 was changed to the original amino acid serine (AGT) by substituting CA with GT, which resulted in three nucleotide changes in the original microRNA172 binding site but retained the original amino acid sequence (Fig. 4a). The vector was transformed into Dongjin plants, which produced the transgenic line DJ-SUI4sd. The transgenic lines showed more severe dwarf phenotypes and much more severely reduced internode lengths than sui4 mutants (Fig. 4b-d). Therefore, the amino acids encoded by the microRNA172 binding site are not related to phenotypic expression. The observation that DJ-SUI4sd plants exhibited a stronger phenotype than sui4 mutants indicates that the strength of the mRNA binding site affinity could determine phenotypic expression. The expression of Os07g0235800 in leaves from 14, 28, and 77 days old plants of Dongjin, sui4, DJ-SUI4, and DJ-SUI4sd plants were measured by qRT-PCR (Additional file 7: Figure S4). Dongjin showed the lowest expression while DJ-SUI4, and DJ-SUI4sd plants showed higher expression than Dongjin and sui4 plants.
Taken together, these results demonstrate that interruption of the affinity of miR172 to the binding site of Os07g0235800 was the cause of reduction in internode length. Interestingly, Os07g0235800 has been reported to be the SNB gene, which controls the transition from spikelet meristem to floral meristem and floral organ development (Lee et al. 2007). Therefore, it was concluded that SUI4 and SNB are the same gene, controlling both internode elongation and floral development during the reproductive stage.
Analysis of the expression pattern of SUI4/SNB using promoter-GUS systems
Since previous work on Os07g0235800 as SNB has focused on floral organs or panicles (Lee et al. 2007), it was necessary to examine the expression of Os07g0235800 during the growth of culms. In order to obtain information on spatial and temporal gene expression patterns, a fusion construct between the 2-kb SUI4/SNB promoter and the GUS reporter gene was introduced into Dongjin plants. In the month prior to the heading stage, when panicles were developed and internodes became elongated, culms of transgenic plants were taken weekly to analyze GUS expression patterns. When panicle length reached 0.6 cm, GUS expression was observed in nodes and the lower parts of internodes where internodes were expected to be elongated (Fig. 5a). In plants with young panicles of 3.5 cm, GUS expression was observed in nodes and the lower part of the uppermost internode. In contrast, little GUS expression was detected in the lower internodes that ceased to elongate (Fig. 5b). When the panicle length reached 12.4 cm, GUS expression was observed in the panicles, nodes, and lower parts of the uppermost internode, but there was little GUS expression in the lower internodes that had ceased elongation (Fig. 5c). When panicle length reached 17.7 cm, GUS expression was observed in nodes and the lower part of elongating upper internodes, but there was little GUS expression in lower internodes (Fig. 5d). During the heading stage, GUS expression was observed in nodes and the lower parts of the two uppermost internodes, but there was little GUS expression in the lower internodes (Fig. 5e).
GUS expression pattern of SNB promoter:GUS fusion gene transgenic lines. (a) Culm of rice plant when panicle length was 0.6 cm. (b) Culm of rice plant when panicle length was 3.5 cm. (c) Culm of rice plant when panicle length was 12.4 cm. (d) Culm of rice plant when panicle length was 17.7 cm. (e) Culm of rice plant when panicle length was 20.8 cm (heading stage). (f) Results of qRT-PCR analysis of the SUI4 gene in the culm of a heading stage plant. I1L: lower half of the first internode, I1U: upper half of the first internode, I2L: lower half of the second internode, I2U: upper half of the second internode, I3L: lower half of the third internode, I3U: upper half of the third internode, N1: first node, N2: second node, N3: third node
Additionally, qRT-PCR was performed to verify the expression of SUI4/SNB in culms in the heading stage. The qRT-PCR data confirmed that expression was higher in upper internodes than in lower internodes, and that within each internode, lower parts showed higher expression than upper parts (Fig. 5f). Overall, SUI4/SNB was expressed in nodes and the lower part of elongating internodes in culms. These expression patterns are consistent with the idea that SUI4/SNB controls internode elongation.
Transcriptome analysis of sui4 mutants vs wild-type Dongjin plants suggests the inactivation of GA and cytokinin
To explore the molecular effects of SUI4/SNB on culm elongation, RNA-seq analysis was carried out with culms in the booting stage. A bioinformatic study was performed to identify differentially expressed genes (DEGs) between sui4 mutants and wild-type (Dongjin) plants. Compared with Dongjin plants, a total of 217 genes were found to be upregulated while 405 genes were downregulated in sui4 mutants (≥ two-fold change; P < 0.05; Additional file 2: Table S2). These DEGs were subjected to gene ontology (GO) enrichment analysis. In terms of biological processes, carbohydrate biosynthesis, polysaccharide biosynthesis, glucan biosynthesis, cellular polysaccharide metabolism, and polysaccharide metabolism were significantly influenced by SUI4/SNB (Table 1). Representative DEGs likely to be related to the sui4 phenotype were selected and their expression in booting stage culms was measured by qRT-PCR (Fig. 6). GIBBERELLIN 2-OXIDASE 3, which inactivates endogenous bioactive gibberellins (GA1, 4) (Lo et al. 2008), was markedly upregulated in sui4 mutants. The depletion of bioactive GAs by increased expression of GIBBERELLIN 2-OXIDASE 3 could be one of the major causes of internode length reduction in sui4 mutants. Also, GIBBERELLIN 20-OXIDASE 1, which produces inactive GA precursors (GA9, 20) (Yamaguchi 2008), was also upregulated. It is reasonable to speculate that the upregulation of GIBBERELLIN 20-OXIDASE 1 might be due to feedback from the decrease of bioactive GAs caused by upregulation of GIBBERELLIN 2-OXIDASE 3. CYTOKININ OXIDASE 9 (OsCKX 9), which inactivates cytokinin (Wang et al. 2014), was also upregulated in sui4 mutants, which might reduce cell division in sui4 internodes. The ONION1 (ONI1) gene which encodes a fatty acid elongase involved in the synthesis of very-long-chain fatty acids and normal shoot development in rice (Ito et al. 2011) was downregulated in sui4 mutants. Also, DIHYDROSPHINGOSINE C4 HYDROXYLASE 1 (DSH1) gene which was reported to mediate biosynthesis of sphingolipid and be abundantly expressed in vascular bundles and apical meristems (Imamura et al. 2007) was downregulated in sui4 mutants. In addition, Os12g0104400 which was annotated as a fatty acid desaturase type 1 domain containing protein gene was downregulated. Although the roles of fatty acid desaturase in cell division and/or cell elongation have not been elucidated fully, this gene might be involved in cell membrane synthesis required for cell expansion. Moreover, Os01g0134500 which was annotated as a delta-7-sterol-C5 desaturase was downregulated. This gene might be involved in brassinosteroid (BR) biosynthesis considering that the Arabidopsis dwarf7 mutant was defective in delta-7-sterol-C5 desaturase gene and BR biosynthesis was hampered in this mutant (Choe et al. 1999). It was noted that OsPHI-1, which has been reported to be involved in cell expansion (Aya et al. 2014), was downregulated. In addition, several genes which may be related to cell wall synthesis and cell expansion were downregulated: Os07g0529700 (xyloglucan endo-transglycosylase-like protein), Os04g0530100 (similar to the beta-expansin 1 precursor AtEXPB1), Os01g0842400 (similar to laccase) and Os09g0262000 (similar to cinnamoyl CoA reductase). Xyloglucan endo-transglycosylase was proposed to do roles of cell wall loosening, assembly, strengthening and expansion (Frankova and Fry 2013), and expansins induce cell wall extension during growth (Li et al. 2003). Laccase and cinnamoyl CoA reductase are involved in biosynthesis of lignin which is predominantly deposited in the secondary cell walls (Park et al. 2017; Swetha et al. 2018). These results indicate that SUI4/SNB activates GA and cytokinin degradation and suppresses genes involved in cell expansion in rice culms.
Relative expression of representative DEGs in the culm of booting stage plants, as revealed by qRT-PCR. RNA was extracted from main culms of sui4 mutant and Dongjin palnts taken at the booting stage. The expression level in Dongjin plants was set as 1. Three biological replicates were performed. Error bars show SE (n = 3)
Modification of flower structure
Previous work investigating the effects SNB on flower structure was performed with knockout mutants (Lee and An 2012; Lee et al. 2007). Our study with sui4 mutants found alterations in floral structure (Additional file 6: Figure S3): wild-type plants have two sterile lemmas, one lemma, and one palea, while the sui4 mutant has one sterile lemma, two lemmas, and one palea. DJ-SUI4 transgenic plants showed a flower structure similar to that of the sui4 mutant. It has been reported that the transition from spikelet meristem to floral meristem was delayed in SNB knockout plants, resulting in the production of multiple rudimentary glumes (Lee et al. 2007). In sui4 mutant and DJ-SUI4 transgenic plants, SNB was upregulated. Therefore, it might be inferred that upregulation of SNB caused a reduction in the number of sterile lemmas and an increase in the number of lemmas.
Discussion
In rice, the majority of the genes determining plant height via internode elongation are directly related to phytohormone GA, BR, or strigolactone biosynthetic or signaling pathways (for review, Liu et al. 2018). However, the present study showed that SUI4/SNB encoding a nuclear protein carrying two AP2 domains (Lee et al. 2007) reduces internode elongation by suppressing cell elongation and cell wall synthesis in the culm. Recently, OsAP2–39 (Oryza sativa APETALA2–39) carrying one AP2 domain was reported to be involved in regulating plant height by controlling the expression of ABA and GA biosynthesis genes (Yaish et al. 2010). Constitutive overexpression of OsAP2–39 under the ubiquitin promoter leads not only to short internodes but also to a low number of tillers and late flowering. Our RNA-seq data strongly suggests that GIBBERELLIN 2-OXIDASE 3, which inactivates endogenous bioactive gibberellins (GA1, 4), and CYTOKININ OXIDASE 9 (OsCKX9), which inactivates cytokinin (Wang et al. 2014), were upregulated in sui4 mutants. Therefore, it is reasonable to state that these AP domain genes operate regulatory mechanisms that control the hormonal homeostasis necessary for internode elongation. Notably, it was reported that the second AP2 domain of Arabidopsis APETALA2 (AP2) which has two AP2 domains like SUI4/SNB binds ‘TTTGTT’ motif and the presence of this motif in the AGAMOUS (AG) second intron is important for the restriction of AG expression in vivo (Dinh et al. 2012). We investigated the presence of ‘TTTGTT’ motif in 2-kb upstream promoter and genic region of the five representative differentially expressed genes which are, GIBBERELLIN 2-OXIDASE 3, GIBBERELLIN 20-OXIDASE 1, OsCKX 9, ONION 1, and OsPHI-1 (Additional file 8: Figure S5). All of these genes have ‘TTTGTT’ motifs in promoter region, which indicates possibility of direct binding of SUI4/SNB to the promoter region. However, further molecular study is required to determine whether these genes are direct targets of SUI4/SNB.
Originally, SUI4/SNB was identified as regulator of flower development (Lee et al. 2007). In contrast to phenotypes induced by knockout mutation, sui4 mutants were able to maintain the native promoter-driven expression pattern by removing the suppression mechanism of microRNA. Instead of the production of multiple rudimentary glumes seen in SNB knockout plants (Lee et al. 2007), sui4 mutants produced one more lemma and one less sterile lemma than wild-type plants.
The most valuable contribution of the present study is the evidence that the microRNA172 regulatory system is involved in the genetic network of internode elongation, which is developmentally controlled by phytohormonal action. The suppression of SNB by microRNA172 has already been well characterized (Lee et al. 2007; Lee et al. 2010; Zhu and Helliwell 2011). The present study showed that SUI4/SNB and microRNA172 play important roles not only in internode elongation but also in floral development. Since elongation of UPIs occurs concomitantly with panicle development, it has been suggested that signaling communication might take place between panicles and UPIs during the reproductive phase (Itoh et al. 2005; Wang et al. 2009; Yamamuro et al. 2000); the present study supports possible signaling pathways that mediate coincidence of panicle development and upper internode elongation. In both dicotyledons and monocotyledons, the expression level of microRNA172 increases as plants grow (Zhu and Helliwell 2011). Furthermore, mature microRNA172 accumulation increased significantly in leaves but not in roots as plants grew, reaching a maximum in the flag leaf (Zhu et al. 2009). It is possible that SNB suppresses internode elongation during the vegetative stage, and that this suppression ceases as microRNA172 increases in the culm during the reproductive stage. Further detailed study of the relationship between SUI4/SNB and microRNA172 expression during rice culm development should provide further insight into the possible SUI4/SNB and microRNA172 regulatory circuit controlling coincident development of panicles and upper internodes.
Conclusion
Due to a mutation at the microRNA172 binding site in sui4 mutants, the expression of SUI4/SNB was enhanced, which lowered the activities of cell expansion and cell wall synthesis and consequently resulted in shortened internodes. This indicates that interaction between SUI4/SNB and microRNA172 could determine internode elongation during the reproductive stage in rice plants.
Materials and methods
Plant materials
Dongjin and sui4 mutant Oryza sativa seeds were sown and the seedlings were grown in a greenhouse for one month before being transplanted into a paddy field. Two- and four-week-old seedlings were taken from the greenhouse and 70-day-old plants were taken from the field and RNA was extracted from the leaves. Additionally, RNA was extracted from the leaves, culms, and panicles of plants grown in the field in the booting and heading stages. At every stage, RNA was extracted from three independent plants. Five plants in the maturing stage were taken for measurement of internode lengths. The middle parts of the uppermost internodes in maturing stage plants were cut and fixed using 10% neutral buffered formalin solution. Then, these samples were embedded in paraffin, sectioned using a microtome, stained with hematoxylin, and observed using an AXIO Imager M1 microscope (ZEISS, Overkochen, Germany). With the photographs, the lengths of cells in parenchyma tissue of the uppermost internode were measured. The cells in three samples were measured for Dongjin plants and sui4 mutants, respectively.
Resequencing
The genomic DNA of sui4 mutant plants was extracted using a DNeasy Plant Maxi Kit (QIAGEN, Hilden, Germany) and used for the preparation of sequencing libraries following the manufacturer’s protocols (Illumina, San Diego, USA). Fragments of the libraries were paired-end sequenced using a HiSeq™ 2000 platform (Illumina, San Diego, USA). The genome sequence data of Dongjin produced using a HiSeq™ 2000 platform was downloaded from Sequence Read Archive (SRA) database of NCBI (https://www.ncbi.nlm.nih.gov/sra): Their accession numbers were ERR157946 and ERR157947. The length of all sequences generated was 101 nucleotides. High-quality raw reads, based on Phred quality values > Q20, were used to analyze genetic variation. The genetic sequence of Oryza sativa L. cv. Nipponbare was used as a reference (Pseudomolecules IRGSP-1.0, https://rapdb.dna.affrc.go.jp/download/irgsp1.html, Rice Genome Sequencing Project 2008). The CLC Assembly Cell program (ver. 3.2.2, http://www.clcbio.com) was used for read mapping and SNP detection.
Candidate gene isolation and transformation
The full Os07g0235800 genomic region from Dongjin plants was amplified by a primer pair of 5′- CGGTACCCGGGGATCCAAATCAGTTCCTACGTGCAACGGGCGCAAAATATC-3′ and 5′- CGACTCTAGAGGATCCGCCCCACTTCCTTGGGAAACTTAAACGATGAGCTC-3′ using PrimeSTAR GXL Polymerase (TaKaRa, Shiga, Japan). The PCR products were purified by gel elution and introduced into a pCAMBIA1300 vector using an In-Fusion HD Cloning Kit (Clontech, Mountain View, USA). The transformation vector thus produced was sequence-verified by sequencing the introduced gene, then used for transformation of sui4 mutants. The full Os07g0235800 genomic region from sui4 mutants was amplified and introduced into the pCAMBIA1300 vector in the same way. The transformation vector thus produced was used for transformation of Dongjin plants. Site-directed mutagenesis was performed with the aforementioned sui4 transformation vector using a QuickChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, USA) according to the instruction manual. A primer pair consisting of 5′- CCATTACTCCCTACTGCAGCAAGTTCAGGATTCTCTACGG-3′ and 5′- CCGTAGAGAATCCTGAACTTGCTGCAGTAGGGAGTAATGG-3′ was used for mutagenesis. The vector thus produced was used for transformation of Dongjin plants. To make a fusion construct between the GUS reporter gene and Os07g0235800 2-kb promoter, the promoter region was amplified first by a primer pair of 5′- AAAAAGCAGGCTAAATCAGTTCCTACGTGCAACGGGCGCAAAATATC-3′ and 5′- AGAAAGCTGGGTACTAACCAACCGCTCTCCCCCGCGC-3′ and then with primers attB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′) and attB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′). The resulting attB-PCR products were cloned by a BP clonase reaction (Invitrogen, Calsbad, USA) into the Gateway pDONR 201 cloning vector (Invitrogen). Subsequently, we used an LR clonase (Invitrogen) reaction to insert the fragment into the destination vector, pMDC163. The vector thus produced was used for transformation of Dongjin plants. Once per week during the month before the heading stage, when panicle development and internode elongation occurred, culms of transgenic plants were taken and the GUS expression pattern was observed. Histochemical GUS staining of transformants was performed according to the protocol described by Dai et al. (1996).
RNA-seq analysis and qRT-PCR
The main culms of sui4 mutant plants and wild-type Dongjin plants were taken at the booting stage. RNA was extracted from culms using an RNeasy Plant Mini Kit (QIAGEN). Three RNA samples from three main culms were used for RNA-seq analysis for sui4 mutant and Dongjin plants, respectively. Total RNA quality and quantity was verified using a NanoDrop1000 spectrometer (Thermo Scientific, Wilmington, USA) and Bioanalyzer 2100 (Agilent technologies). We used TruSeq RNA Library Preparation Kit (Illumina) for sequencing library preparation. The TruSeq library was sequenced using the HiSeq™ 2000 platform (Illumina). The RNA-seq reads were mapped to the Oryza sativa reference genome using TopHat (Trapnell et al. 2009). The transcript counts were calculated and the relative transcript abundances were measured in FPKM (fragments per kilobase of exon per million fragments mapped) using Cufflinks (Trapnell et al. 2010). For each transcript, we conducted independent t-tests between sui4 mutant and Dongjin plants. Finally, we determined the DEGs by adjusting |fold change| ≥ 2 and independent t-test raw p < 0.05. Biological gene functional annotation analysis was performed for the DEG list using the DAVID tool (http://david.abcc.ncifcrf.gov/) (Huang et al. 2007) to understand the biological meanings behind the list of DEGs. In the DAVID annotation system, modified Fisher’s exact p-values (EASE scores) are adopted to measure gene enrichment in annotation terms. The EASE score was corrected using the Benjamini method to reduce false positives caused by multiple testing. All GO terms < 0.05 after Benjamini correction were taken as significantly enriched GO terms.
qRT-PCR analysis for the representative DEGs was performed using the RNA used for RNA-seq. cDNA was synthesized using a PrimeScript 1st strand cDNA Synthesis Kit (Takara). Primers, shown in Additional file 3: Table S3, were designed on the GenScript Real-time PCR Primer Design website (https://www.genscript.com/ssl-bin/app/primer). The rice ubiquitin 1 gene was used as a reference gene for normalization. PCR amplifications were performed in 20 μl final volumes containing AccuPower 2X GreenStar qPCR MasterMix (BiONEER, Daejeon, Korea), 10 pmol of each primer, and 2 μl cDNA solution on a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, USA). Three biological replicates were performed. Relative gene expression was calculated by Bio-Rad CFX Manager 3.1 software (Bio-Rad).
Accession codes
The sequencing data of sui4 mutant have been submitted to NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number of SRR7942370 and SRR7942369. The accession numbers of the sequencing data of Dongjin were ERR157946 and ERR157947.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files. The accession codes of the resequencing data of sui4 mutant and Dongjin were indicated.
Abbreviations
- BRs:
-
Brassinosteroids
- BZR1 :
-
BRASSINAZOLE-RESISTANT 1
- DEGs:
-
differentially expressed genes
- DZ:
-
differentiation zone
- Ehd1 :
-
EARLY HEADING DATE 1
- EZ:
-
elongation zone
- FPKM:
-
fragments per kilobase of exon per million fragments mapped
- GA:
-
Gibberellin
- GO:
-
Gene ontology
- GUS:
-
β-glucuronidase
- IBH1 :
-
ILI1 BINDING BHLH 1
- ILI1 :
-
INCREASED LAMINA INCLINATION 1
- IM:
-
intercalary meristem
- miR172:
-
microRNA172
- ONI1 :
-
ONION1
- OsAP2–39 :
-
Oryza sativa APETALA2–39
- OsCKX 9 :
-
Oryza sativa CYTOKININ OXIDASE 9
- OsGA20OX1 :
-
Oryza sativa GIBBERELLIN 20-OXIDASE 1
- OsGA2OX3 :
-
Oryza sativa GIBBERELLIN 2-OXIDASE 3
- OsIDS1 :
-
Oryza sativa INDETERMINATE SPIKELET1
- PRE1 :
-
PACLOBUTRAZOL RESISTANCE 1
- qRTPCR:
-
quantitative reverse transcription-PCR
- SLR 1 :
-
SLENDER RICE1
- SNB :
-
SUPERNUMERARY BRACT
- sui4 :
-
shortened uppermost internode 4
- UPIs:
-
Upper internodes
References
Aya K, Hobo T, Sato-Izawa K, Ueguchi-Tanaka M, Kitano H, Matsuoka M (2014) A novel AP2-type transcription factor, SMALL ORGAN SIZE1, controls organ SIZE downstream of an auxin signaling pathway. Plant Cell Physiol 55(5):897–912. https://doi.org/10.1093/pcp/pcu023
Choe S, Noguchi T, Fujioka S, Takatsuto S, Tissier CP, Gregory BD, Ross AS, Tanaka A, Yoshida S, Tax FE, Feldmann KA (1999) The Arabidopsis dwf7/ste1 mutant is defective in the delta7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 11(2):207–221
Chonan N (1993) Chapter 3. Stem. In: Matsuo T, Hoshikawa K (eds) Science of the Rice Plant, vol 1. Nobunkyo, Tokyo, pp 187–188
Dai Z, Gao J, An K, Lee JM, Edwards GE, An G (1996) Promoter elements controlling developmental and environmental regulation of a tobacco ribosomal protein gene L34. Plant Mol Biol 32(6):1055–1065
Dinh TT, Girke T, Liu X, Yant L, Schmid M, Chen X (2012) The floral homeotic protein APETALA2 recognizes and acts through an AT-rich sequence element. Development 139:1978–1986. https://doi.org/10.1242/dev.077073
Frankova L, Fry SC (2013) Biochemistry and physiological roles of enzymes that 'cut and paste' plant cell-wall polysaccharides. J Exp Bot 64(12):3519–3550. https://doi.org/10.1093/jxb/ert201
Huang D, Wang S, Zhang B, Shang-Guan K, Shi Y, Zhang D, Liu X, Wu K, Xu Z, Fu X, Zhou Y (2015) A gibberellin-mediated DELLA-NAC signaling Cascade regulates cellulose synthesis in Rice. Plant Cell 27(6):1681–1696. https://doi.org/10.1105/tpc.15.00015
Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA (2007) DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic acids res 35(web server issue):W169-175. https://doi.org/10.1093/nar/gkm415
Imamura T, Kusano H, Kajigaya Y, Ichikawa M, Shimada H (2007) A rice dihydrosphingosine C4 hydroxylase (DSH1) gene, which is abundantly expressed in the stigmas, vascular cells and apical meristem, may be involved in fertility. Plant Cell Physiol 48(8):1108–1120. https://doi.org/10.1093/pcp/pcm084
Ito Y, Kimura F, Hirakata K, Tsuda K, Takasugi T, Eiguchi M, Nakagawa K, Kurata N (2011) Fatty acid elongase is required for shoot development in rice. Plant J 66(4):680–688. https://doi.org/10.1111/j.1365-313X.2011.04530.x
Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46(1):23–47
Je BI, Piao HL, Park SJ, Park SH, Kim CM, Xuan YH, Park SH, Huang J, Do Choi Y, An G, Wong HL, Fujioka S, Kim MC, Shimamoto K, Han CD (2010) RAV-Like1 maintains brassinosteroid homeostasis via the coordinated activation of BRI1 and biosynthetic genes in rice. Plant Cell 22(6):1777–1791
Ji H, Kim H, Yun D-W, Yoon U-H, Kim T-H, Eun M-Y, Lee G-S (2014) Characterization and fine mapping of a shortened uppermost internode mutant in rice. Plant biotechnology reports 8:125-134. Doi. https://doi.org/10.1007/s11816-013-0280-5
Kende H, van der Knaap E, Cho HT (1998) Deepwater rice: a model plant to study stem elongation. Plant Physiol 118(4):1105–1110
Lee DY, An G (2012) Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J 69(3):445–461. https://doi.org/10.1111/j.1365-313X.2011.04804.x
Lee DY, Lee J, Moon S, Park SY, An G (2007) The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J 49(1):64–78. https://doi.org/10.1111/j.1365-313X.2006.02941.x
Lee H, Yoo SJ, Lee JH, Kim W, Yoo SK, Fitzgerald H, Carrington JC, Ahn JH (2010) Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res 38(9):3081–3093. https://doi.org/10.1093/nar/gkp1240
Lee YS, Lee DY, Cho LH, An G (2014) Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice (N Y) 7(1):31. https://doi.org/10.1186/s12284-014-0031-4
Li Y, Jones L, McQueen-Mason S (2003) Expansins and cell growth. Curr Opin Plant Biol 6(6):603–610
Liu F, Wang P, Zhang X, Li X, Yan X, Fu D, Wu G (2018) The genetic and molecular basis of crop height based on a rice model. Planta 247(1):1–26. https://doi.org/10.1007/s00425-017-2798-1
Lo SF, Yang SY, Chen KT, Hsing YI, Zeevaart JA, Chen LJ, Yu SM (2008) A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20(10):2603–2618. https://doi.org/10.1105/tpc.108.060913
Park HL, Bhoo SH, Kwon M, Lee SW, Cho MH (2017) Biochemical and expression analyses of the Rice Cinnamoyl-CoA reductase gene family. Front Plant Sci 8:2099. https://doi.org/10.3389/fpls.2017.02099
Swetha C, Basu D, Pachamuthu K, Tirumalai V, Nair A, Prasad M, Shivaprasad PV (2018) Major domestication-related phenotypes in Indica Rice are due to loss of miRNA-mediated laccase silencing. Plant Cell 30(11):2649–2662. https://doi.org/10.1105/tpc.18.00472
Teotia S, Tang G (2015) To bloom or not to bloom: role of microRNAs in plant flowering. Mol Plant 8(3):359–377. https://doi.org/10.1016/j.molp.2014.12.018
Tong H, Xiao Y, Liu D, Gao S, Liu L, Yin Y, Jin Y, Qian Q, Chu C (2014) Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26(11):4376–4393. https://doi.org/10.1105/tpc.114.132092
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111. https://doi.org/10.1093/bioinformatics/btp120
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515. https://doi.org/10.1038/nbt.1621
Wang L, Yin H, Qian Q, Yang J, Huang C, Hu X, Luo D (2009) NECK LEAF 1, a GATA type transcription factor, modulates organogenesis by regulating the expression of multiple regulatory genes during reproductive development in rice. Cell Res 19(5):598–611
Wang W, Li G, Zhao J, Chu H, Lin W, Zhang D, Wang Z, Liang W (2014) Dwarf Tiller1, a Wuschel-related homeobox transcription factor, is required for tiller growth in rice. PLoS Genet 10(3):e1004154. https://doi.org/10.1371/journal.pgen.1004154
Yaish MW, El-Kereamy A, Zhu T, Beatty PH, Good AG, Bi YM, Rothstein SJ (2010) The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet 6(9):e1001098. https://doi.org/10.1371/journal.pgen.1001098
Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59:225–251
Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12(9):1591–1606
Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, Yang H, Xu Y, Kim SH, Fujioka S, Lin WH, Chong K, Lu T, Wang ZY (2009) Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21(12):3767–3780. https://doi.org/10.1105/tpc.109.070441
Zhu QH, Helliwell CA (2011) Regulation of flowering time and floral patterning by miR172. J Exp Bot 62(2):487–495. https://doi.org/10.1093/jxb/erq295
Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA (2009) Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol 9:149. https://doi.org/10.1186/1471-2229-9-149
Acknowledgements
We thank Hee-Ja Kim for her rice transformation works.
Funding
This research was supported by grants from the National Institute of Agricultural Sciences (NAS), project code PJ01246802, Republic of Korea.
Author information
Authors and Affiliations
Contributions
HJ and CDH conceived of the study, participated in its design, and wrote the paper. GSL did field works and phenotyping. KHJ performed RNA-seq analysis. DYK, JO, HO, and SLK performed DNA and RNA extraction, gene cloning, and gene expression analysis. KSC maintained transgenic plant materials in greenhouse. ICH and JB performed genome sequence data analysis. KHK provided an overall direction for this project.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. List of SNPs in the SUI4 mapped region of Dongjin plants and sui4 mutants. (DOCX 16 kb)
Additional file 2:
Table S2. Differentially expressed genes (at least two fold change in expression, p-value< 0.05) in culm (XLSX 55 kb)
Additional file 3:
Table S3. qRT-PCR primer sequences (DOCX 14 kb)
Additional file 4:
Figure S1. RT-PCR of candidate genes for the sui4 mutant phenotype. Expression of the five genes with SNPs between Dongjin and sui4 mutant in the mapped region was measured by RT-PCR. (JPG 20 kb)
Additional file 5:
Figure S2. Culm structure of Os07g0235800 transgenic plants. Arrows indicate locations of nodes. Two main culms from two plants are shown for each phenotype. Two independent transgenic lines are shown for SUI4-DJ and DJ-SUI4. (JPG 49 kb)
Additional file 6:
Figure S3. Flower structure of an sui4 mutant and a transgenic line. (A), (B): wild type, (C), (D): sui4 mutant, (E), (F): DJ-SUI4 transgenic line. rg: rudimentary glume, sl: sterile lemma, le: lemma, pa: palea (JPG 73 kb)
Additional file 7:
Figure S4. Os07g0235800 gene expression in Dongjin, sui4, DJ-SUI4, and DJ-SUI4sd plants. Relative fold expression difference is based on the expression level detected in 14 day old seedling leaves of Dongjin. Error bars represent standard deviation of the expression ratio. 14 L: 14 day old seedling leaves, 28 L: 28 day old seedling leaves, 77 L: 77 day old plant leaves, DJ: Dongjin, DJ-SUI4: transgenic line in which the SUI4 gene from a sui4 mutant was introduced into Dongjin plants, DJ-SUI4sd: transgenic line in which the SUI4 gene, modified by site-directed mutagenesis as shown in Fig. 3a, was introduced into Dongjin plants. (JPG 42 kb)
Additional file 8:
Figure S5. Location of AP2 domain binding motif ‘TTTGTT’ in promoter and genic regions in the five representative genes which are differentially expressed between Dongjin plants and sui4 mutants. The locations of ‘TTTGTT’ motif are indicated by brown vertical lines. Filled gray boxes indicate 5′ and 3′ UTRs, and filled black boxes indicate exons, including protein-coding sequence. Gray lines indicate introns, and green lines indicate 2-kb upstream promoter region. (JPG 39 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ji, H., Han, Cd., Lee, GS. et al. Mutations in the microRNA172 binding site of SUPERNUMERARY BRACT (SNB) suppress internode elongation in rice. Rice 12, 62 (2019). https://doi.org/10.1186/s12284-019-0324-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-019-0324-8