Abstract
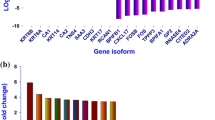
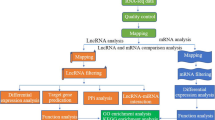
Long non-coding RNA (lncRNA) was previously considered as a non-functional transcript, which now established as part of regulatory elements of biological events such as chromosome structure, remodeling, and regulation of gene expression. The study presented here showed the role of lncRNA through differential expression analysis on cancer-related coding genes in horn squamous cell carcinoma of Indian zebu cattle. A total of 10,360 candidate lncRNAs were identified and further analyzed for its coding potential ability using three tools (CPC, CPAT, and PLEK) that provide 8862 common lncRNAs. Pfam analysis of these common lncRNAs gave 8612 potential candidates for lncRNA differential expression analysis. Differential expression analysis showed a total of 59 significantly differentially expressed genes and 19 lncRNAs. Pearson’s correlation analysis was used to identify co-expressed mRNA-lncRNAs to established relation of the regulatory role of lncRNAs in horn cancer. We established a positive relation of seven upregulated (XLOC_000016, XLOC_002198, XLOC_002851, XLOC_ 007383, XLOC_010701, XLOC_010272, and XLOC_011517) and one downregulated (XLOC_011302) lncRNAs with eleven genes that are related to keratin family protein, keratin-associated protein family, cornifelin, corneodesmosin, serpin family protein, and metallothionein that have well-established role in squamous cell carcinoma through cellular communication, cell growth, cell invasion, and cell migration. These biological events were found to be related to the MAPK pathway of cell cycle regulation indicating the role of lncRNAs in manipulating cell cycle regulation during horn squamous cell carcinomas that will be useful in identifying molecular portraits related to the development of horn cancer.







Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article as supplementary information files. In addition, the datasets generated and analyzed during the current study are available in the NCBI-SRA with bioproject number PRJNA497667 and biosample numbers as given in supplementary data. The following link can be used by the reviewer to access data as a part of data share at NCBI-SRA that are publically available. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA497667
Abbreviations
- HN:
-
horn normal
- HC:
-
horn cancer
- CPC:
-
Coding Potential Calculator
- CPAT:
-
Coding-Potential Assessment Tools
- PLEK:
-
predictor of long non-coding RNAs and messenger RNAs based on an improved K-mer scheme
- lncRNA:
-
long non-coding RNA
- FPKM:
-
fragments per kilobase of transcripts per millions mapped reads
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
References
Asadzadeh-Aghdaei H, Zali MR, Bonad MA et al (2018) The application of gene expression profiling in predictions of occult lymph node metastasis in colorectal cancer patients. Biomedicines 6:27
Baruzzo G, Hayer KE, Kim EJ, Di Camillo B, FitzGerald GA, Grant GR (2017) Simulation-based comprehensive benchmarking of RNA-seq aligners. Nat Methods 14:135–139
Batista PJ, Chang HY (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell 152:1298–1307
Benesty J, Chen J, Huang Y, Cohen I (2009) Pearson correlation coefficient. In: Noise reduction in speech processing. Springer, Springer Topics in Signal Processing, vol 2. Springer, Berlin, Heidelberg, 37–40. https://doi.org/10.1007/978-3-642-00296-0_5
Billerey C, Boussaha M, Esquerre D et al (2014) Identification of large intergenic non-coding RNAs in bovine muscle using next-generation transcriptomic sequencing. BMC Genomics 15:499
Boldrup L, Gu X, Coates PJ et al (2017) Gene expression changes in tumor free tongue tissue adjacent to tongue squamous cell carcinoma. Oncotarget 8:19389
Burggraaf H (1935) Kanker aan de basis van de hoorns bij zebus, Horn-core disease of cattle. T Diergeneesk 62:1121-1136
Caballero J, Gilbert I, Fournier E, Gagné D, Scantland S, Macaulay A, Robert C (2015) Exploring the function of long non-coding RNA in the development of bovine early embryos. Reprod Fertil Dev 27:40–52
Chattopadyay S, Jandrotia V, Iyer P (1982) Horn cancer in sheep. Indian Veterinary Journal 59:319–320
Chidambaranathan Reghupaty S (2017) TAF2: a potential oncogene for hepatocellular carcinoma. Dessertation, Virginia Commonwealth University
Chiu H-S, Somvanshi S, Patel E et al (2018) Pan-cancer analysis of lncRNA regulation supports their targeting of cancer genes in each tumor context. Cell Rep 23:297–312.e12
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Consortium IHGS (2004) Finishing the euchromatic sequence of the human genome. Nature 431:931
Dalla Pozza E, Manfredi M, Brandi J, Buzzi A, Conte E, Pacchiana R, Cecconi D, Marengo E, Donadelli M (2018) Trichostatin A alters cytoskeleton and energy metabolism of pancreatic adenocarcinoma cells: an in depth proteomic study. J Cell Biochem 119:2696–2707
Damodaran S, Sundararaj A, Ramakrishnan R (1979) Horn cancer in bulls. The Indian Veterinary Journal 56:248–249
Dankert JT, Wiesehöfer M, Czyrnik ED, Singer BB, von Ostau N, Wennemuth G (2018) The deregulation of miR-17/CCND1 axis during neuroendocrine transdifferentiation of LNCaP prostate cancer cells. PLoS One 13:e0200472
de Koning PJ, Kummer JA, de Poot SAH et al (2011) Intracellular serine protease inhibitor SERPINB4 inhibits granzyme M-induced cell death. PLoS One 6:e22645
Do DN, Ibeagha-Awemu EM (2017) Non-coding RNA roles in ruminant mammary gland development and lactation. In: Current Topics in Lactation. InTech Open, 5. https://doi.org/10.5772/67194
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21
Dong D, Mu Z, Zhao C, Sun M (2018) ZFAS1: a novel tumor-related long non-coding RNA. Cancer Cell Int 18:125
Dziegiel P, Pula B, Kobierzycki C, Stasiolek M, Podhorska-Okolow M (2016) The role of metallothioneins in carcinogenesis Advances in Anatomy, Embryology and Cell Biology 218:1–117
Escalona M, Rocha S, Posada D (2016) A comparison of tools for the simulation of genomic next-generation sequencing data. Nat Rev Genet 17:459–469
Flynn RA, Chang HY (2014) Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 14:752–761
Ghosh S (2018) Epitopes of enzymes involved in sialylation with special reference to lung cancer. Research & reviews: a journal of life sciences 8:11–19
Gładych M, Cylwa R, Kiełczewski K, Biecek P, Liloglou T, Mackiewicz A, Oleksiewicz U (2018) The expression signature of cancer-associated KRAB-ZNF factors identified in TCGA pan-cancer transcriptomic data. Mol Oncol 13:701–724
Goff L, Trapnell C, Kelley D (2013) cummeRbund: Analysis, exploration, manipulation, and visualization of Cufflinks high-throughput sequencing data Rpackage version 2.26.0
Gupta R, Sadana J, Kuchroo V, Kalra D (1980) Horn cancer in an intact bull. The Veterinary Record 107:312
Ho DW-H, Kai AK-L, Ng IO-L (2015) TCGA whole-transcriptome sequencing data reveals significantly dysregulated genes and signaling pathways in hepatocellular carcinoma. Front Med 9:322–330
Hon C-C, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJL, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, Lizio M, Kawaji H, Kasukawa T, Itoh M, Burroughs AM, Noma S, Djebali S, Alam T, Medvedeva YA, Testa AC, Lipovich L, Yip CW, Abugessaisa I, Mendez M, Hasegawa A, Tang D, Lassmann T, Heutink P, Babina M, Wells CA, Kojima S, Nakamura Y, Suzuki H, Daub CO, de Hoon MJL, Arner E, Hayashizaki Y, Carninci P, Forrest ARR (2017) An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543:199–204
Hu J, Gao Y, Zheng Y, Shang X (2018a) KF-finder: identification of key factors from host-microbial networks in cervical cancer. BMC Syst Biol 12:54
Hu X, Zhai Y, Shi R et al (2018b) FAT1 inhibits cell migration and invasion by affecting cellular mechanical properties in esophageal squamous cell carcinoma. Oncol Rep 39:2136–2146
Huang G-J, Luo M-S, Chen G-P, Fu M-Y (2018) MiRNA–mRNA crosstalk in laryngeal squamous cell carcinoma based on the TCGA database. Eur Arch Otorhinolaryngol 275:751–759
Ibeagha-Awemu EM, Do DN, Dudemaine P-L, Fomenky BE, Bissonnette N (2018) Integration of lncRNA and mRNA transcriptome analyses reveals genes and pathways potentially involved in calf intestinal growth and development during the early weeks of life. Genes 9:142
Izuhara K, Yamaguchi Y, Ohta S, Nunomura S, Nanri Y, Azuma Y, Nomura N, Noguchi Y, Aihara M (2018) Squamous cell carcinoma antigen 2 (SCCA2, SERPINB4): an emerging biomarker for skin inflammatory diseases. Int J Mol Sci 19:1102
Joshi B, Soni P, Fefar D, Ghodasara D, Prajapati K (2009) Epidemiological and pathological aspects of horn cancer in cattle of Gujarat. Indian J Field Vet 5:15–18
Kanehisa M The KEGG database. In: ‘In Silico’ simulation of biological processes: Novartis Foundation Symposium 247, 2002. Wiley Online Library, pp 91–103
Karunanithi S, Levi L, DeVechhio J et al (2017) RBP4-STRA6 pathway drives cancer stem cell maintenance and mediates high-fat diet-induced colon carcinogenesis. Stem Cell Rep 9:438–450
Kohl M, Wiese S, Warscheid B (2011) Cytoscape: software for visualization and analysis of biological networks. In: Data Mining in Proteomics. Springer, pp 291–303
Kong L, Zhang Y, Ye Z-Q, Liu X-Q, Zhao S-Q, Wei L, Gao G (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35:W345–W349
Koringa PG, Jakhesara SJ, Bhatt VD, Patel AB, Dash D, Joshi CG (2013) Transcriptome analysis and SNP identification in SCC of horn in (Bos indicus) Indian cattle. Gene 530:119–126
Koufariotis LT, Chen Y-PP, Chamberlain A, Vander Jagt C, Hayes BJ (2015) A catalogue of novel bovine long noncoding RNA across 18 tissues. PLoS One 10:e0141225
Kulkarni H (1953) Carcinoma of horn in bovines of Old Baroda State. Indian Vet J 29:415–421
Kung JT, Colognori D, Lee JT (2013) Long noncoding RNAs: past, present, and future. Genetics 193:651–669
Lamontagne S, Fortier A-M, Parent S, Asselin E, Cadrin M (2015) Interaction between keratin intermediate filament proteins K8/18 and cancer related signal transduction proteins in epithelial cells. AACR. https://doi.org/10.1158/1538-7445
Lee J, Ngeow J (2018) Inherited thyroid cancer. In: Evidence-Based Endocrine Surgery. Springer, pp 163–171
Lee CW, Lin SE, Tsai HI, Su PJ, Hsieh CH, Kuo YC, Sung CM, Lin CY, Tsai CN, Yu MC (2018) Cadherin 17 is related to recurrence and poor prognosis of cytokeratin 19-positive hepatocellular carcinoma. Oncol Lett 15:559–567
Li A, Zhang J, Zhou Z (2014) PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinf 15:311
Liu X, Ding X, Li X, Jin C, Yue Y, Li G, Guo H (2017) An atlas and analysis of bovine skeletal muscle long noncoding RNAs. Anim Genet 48:278–286
Liu Z, Ye Q, Wu L, Gao F, Xie H, Zhou L, Zheng S, Xu X (2018) Metallothionein 1 family profiling identifies MT1X as a tumor suppressor involved in the progression and metastastatic capacity of hepatocellular carcinoma. Mol Carcinog 57:1435–1444
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408
Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ (2012) Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30:434–439
Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448–3449
Marques AC, Ponting CP (2009) Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol 10:R124
Mazzoccoli G, Castellana S, Carella M et al (2017) A primary tumor gene expression signature identifies a crucial role played by tumor stroma myofibroblasts in lymph node involvement in oral squamous cell carcinoma. Oncotarget 8:104913
Nie Y, Li S, Zheng X, Chen W, Li X, Liu Z, Hu Y, Qiao H, Qi Q, Pei Q, Cai D, Yu M, Mou C (2018) Transcriptome reveals long non-coding RNAs and mRNAs involved in primary wool follicle induction in carpet sheep fetal skin. Front Physiol 9:446
Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S, Östensson M, Akyürek LM, Abrahamsson J, Pfeifer S, Larsson E, Shi L, Peng Z, Fischer M, Martinsson T, Hedborg F, Kogner P, Kanduri C (2014) The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell 26:722–737
Papa F, Siciliano RA, Inchingolo F, Mazzeo MF, Scacco S, Lippolis R (2018) Proteomics pattern associated with gingival oral squamous cell carcinoma and epulis: a case analysis. Oral Sci Int 15:41–47
Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11:1650–1667
Quail MA, Smith M, Coupland P et al (2012) A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13:341
Rezende A, Naves P (1975) Horn core cancer in a zebu cow, imported to Brazil. Pesq Agropec Bras Série Veterinária 10:41–44
Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145–166
Robles A, Ryan B (2015) KRT81 miR-SNP rs3660 is associated with risk and survival of NSCLC. Ann Oncol 27:360–361
Rong C, Meinert É, Hess J (2018) Estrogen receptor signaling in radiotherapy: from molecular mechanisms to clinical studies. Int J Mol Sci 19:713
Roth L, Srivastava S, Lindzen M et al (2018) SILAC identifies LAD1 as a filamin-binding regulator of actin dynamics in response to EGF and a marker of aggressive breast tumors. Sci Signal 11:eaan0949
Savita J, Kumar BY, Nayak VN (2018) Matrix metalloproteinases in oral squamous cell carcinoma-a review. J Adv Clin Res Insights 5:124–126
Schmitt AM, Chang HY (2016) Long noncoding RNAs in cancer pathways. Cancer Cell 29:452–463
Shi J, Yan B, Lou X, Ma H, Ruan S (2017) Comparative transcriptome analysis reveals the transcriptional alterations in heat-resistant and heat-sensitive sweet maize (Zea mays L.) varieties under heat stress. BMC Plant Biol 17:26
Shiba D, Terayama M, Yamada K, Hagiwara T, Oyama C, Tamura-Nakano M, Igari T, Yokoi C, Soma D, Nohara K, Yamashita S, Dohi T, Kawamura YI (2018) Clinicopathological significance of cystatin A expression in progression of esophageal squamous cell carcinoma. Medicine 97:e0357
Silveira NJ, Varuzza L, Machado-Lima A et al (2008) Searching for molecular markers in head and neck squamous cell carcinomas (HNSCC) by statistical and bioinformatic analysis of larynx-derived SAGE libraries. BMC Med Genet 1:56
Szklarczyk D, Franceschini A, Wyder S et al (2014) STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452
Taft RJ, Pheasant M, Mattick JS (2007) The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays 29:288–299
Tang Q, Zhang H, Kong M, Mao X, Cao X (2018) Hub genes and key pathways of non-small lung cancer identified using bioinformatics. Oncol Lett 16:2344–2354
Teschendorff AE, Lee SH, Jones A, Fiegl H, Kalwa M, Wagner W, Chindera K, Evans I, Dubeau L, Orjalo A, Horlings HM, Niederreiter L, Kaser A, Yang W, Goode EL, Fridley BL, Jenner RG, Berns EMJJ, Wik E, Salvesen HB, Wisman GBA, van der Zee AGJ, Davidson B, Trope CG, Lambrechts S, Vergote I, Calvert H, Jacobs IJ, Widschwendter M (2015) HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med 7:108
Toivola DM, Boor P, Alam C, Strnad P (2015) Keratins in health and disease. Curr Opin Cell Biol 32:73–81
Toss A, Cristofanilli M (2015) Molecular characterization and targeted therapeutic approaches in breast cancer. Breast Cancer Res 17:60
Trapnell C, Robert A, Goff L et al (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578
Tsuchiya KD, Davis B, Gardner RA (2017) Is intrachromosomal amplification of chromosome 21 (iAMP21) always intrachromosomal? Cancer Genet 218:10–14
Vadakekolathu J, Al-Juboon SIK, Johnson C et al (2018) MTSS1 and SCAMP1 cooperate to prevent invasion in breast cancer. Cell Death Dis 9:344
Wang Y, Jin L (2018) miRNA-145 is associated with spontaneous hypertension by targeting SLC7A1. Exp Ther Med 15:548–552
Wang L, Park HJ, Dasari S, Wang S, Kocher J-P, Li W (2013) CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res 41:e74–e74
Weikard R, Hadlich F, Kuehn C (2013) Identification of novel transcripts and noncoding RNAs in bovine skin by deep next generation sequencing. BMC Genomics 14:789
Wickham H (2010) ggplot2: elegant graphics for data analysis. J Stat Softw 35:65–88
Xiong Z, Ren S, Chen H, Liu Y, Huang C, Zhang YL, Odera JO, Chen T, Kist R, Peters H, Garman K, Sun Z, Chen X (2018) PAX9 regulates squamous cell differentiation and carcinogenesis in the oro-oesophageal epithelium. J Pathol 244:164–175
Yang B, Jiao B, Ge W, Zhang X, Wang S, Zhao H, Wang X (2018) Transcriptome sequencing to detect the potential role of long non-coding RNAs in bovine mammary gland during the dry and lactation period. BMC Genomics 19:605
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinf 13:134
Zhang X, Roger G, Ali M et al (2010) Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. https://doi.org/10.1158/0008-5472.CAN-09-3885
Zhang J, Le TD, Liu L, Li J (2018) Inferring and analyzing module-specific lncRNA–mRNA causal regulatory networks in human cancer. Brief Bioinform. https://doi.org/10.1093/bib/bby008
Zheng Y, Zhao G, Xu B, Liu C, Li C, Zhang X, Chang X (2016) PADI4 has genetic susceptibility to gastric carcinoma and upregulates CXCR2, KRT14 and TNF-α expression levels. Oncotarget 7:62159
Zheng X, Ning C, Zhao P et al (2018) Integrated analysis of long noncoding RNA and mRNA expression profiles reveals the potential role of long noncoding RNA in different bovine lactation stages. J Dairy Sci 101:11062–11073
Zombori T, Cserni G (2018) Immunohistochemical analysis of the expression of breast markers in basal-like breast carcinomas defined as triple negative cancers expressing keratin 5. Pathol Oncol Res 24:259–267
Zubaidy AJ (1976) Horn cancer in cattle in Iraq. Vet Pathol 13:453–454
Funding
We thank the Department of Biotechnology (DBT), Government of India, New Delhi, India, for providing financial support (Grant Letter No. BT/PR13649/AAQ/1/627/2015) for this project.
Author information
Authors and Affiliations
Contributions
PHS and KJP participated in sample collection, library preparation, and sequencing; PHS analyzed data and drafted manuscript; PGK conceptualized actual research project, participated in sample collection, and improved manuscript; SJJ helped in data analysis and improved manuscript; CGJ provided all facilities to carryout research. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
All animal ethics guidelines were followed and complied as per permission from Ethical Committee norms and letter No. IAEC 525-2015.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Figure S1
Heatmap showing expression level based upon FPKM value for A) Genes and B) LncRNAs. (PNG 348 kb)
Figure S2
Protein-protein interaction analysis of significant differentially expressed genes. (PNG 105 kb)
Figure S3
Gene ontology analysis based upon significant differentially expressed genes. (PNG 220 kb)
Figure S4
Interaction between different gene ontology terms identified based upon significant differentially expressed genes. (PNG 303 kb)
Figure S5
Interaction between different gene ontology terms identified based upon target genes of lncRNAs. (PNG 249 kb)
Figure S6
Comparison of RT-qPCR and transcriptome analysis results of selected genes and lncRNAs based upon their log2 fold change value. (PNG 86 kb)
ESM 1
(XLSX 1092 kb)
Rights and permissions
About this article
Cite this article
Sabara, P.H., Jakhesara, S.J., Panchal, K.J. et al. Transcriptomic analysis to affirm the regulatory role of long non-coding RNA in horn cancer of Indian zebu cattle breed Kankrej (Bos indicus). Funct Integr Genomics 20, 75–87 (2020). https://doi.org/10.1007/s10142-019-00700-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-019-00700-4