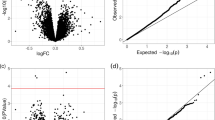
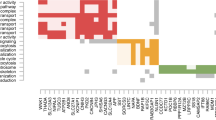
Abstract
Autism was a spectrum of multiple complex diseases that required an interdisciplinary group of experts to make a diagnostic decision. Both genetic and environmental factors play essential roles in causing the onset of Autism. Therefore, this study hypothesized that methylomic biomarkers may facilitate the accurate Autism detection. A comprehensive series of biomarker detection algorithms were utilized to find the best methylomic biomarkers for the Autism detection using the methylomic data of the peripheral blood samples. The best model achieved 99.70% in accuracy with 678 methylomic biomarkers and a tenfold cross validation strategy. Some of the methylomic biomarkers were experimentally confirmed to be associated with the onset or development of Autism.





Similar content being viewed by others
References
Uljarevic M, Lane A, Kelly A, Leekam S (2016) Sensory subtypes and anxiety in older children and adolescents with autism spectrum disorder. Autism Res 9:1073–1078
Yoshimura RF, Tran MB, Hogenkamp DJ, Ayala NL, Johnstone T, Dunnigan AJ, Gee TK, Gee KW (2017) Allosteric modulation of nicotinic and GABAA receptor subtypes differentially modify autism-like behaviors in the BTBR mouse model. Neuropharmacology 126:38–47
Oblak A, Gibbs TT, Blatt GJ (2013) Reduced serotonin receptor subtypes in a limbic and a neocortical region in autism. Autism Res 6:571–583
Deckmann I, Schwingel GB, Fontes-Dutra M, Bambini-Junior V, Gottfried C (2018) Neuroimmune alterations in autism: a translational analysis focusing on the animal model of autism induced by prenatal exposure to valproic acid. Neuroimmunomodulation 25(5–6):285–299
Zaboski BA, Storch EA (2018) Comorbid autism spectrum disorder and anxiety disorders: a brief review. Future Neurol 13:31–37
Lussier AA, Weinberg J, Kobor MS (2017) Epigenetics studies of fetal alcohol spectrum disorder: where are we now? Epigenomics 9:291–311
Shi L, Zhang X, Golhar R, Otieno FG, He M, Hou C, Kim C, Keating B, Lyon GJ, Wang K, Hakonarson H (2013) Whole-genome sequencing in an autism multiplex family. Mol Autism 4:8
Margolis KG, Buie TM, Turner JB, Silberman AE, Feldman JF, Murray KF, McSwiggan-Hardin M, Levy J, Bauman ML, Veenstra-VanderWeele J et al (2018) Development of a brief parent-report screen for common gastrointestinal disorders in autism spectrum disorder. J Autism Dev Disord 49(1):349–362
Fowlie G, Cohen N, Ming X (2018) The perturbance of microbiome and gut-brain axis in autism spectrum disorders. Int J Mol Sci. https://doi.org/10.3390/ijms19082251
Ward J, Hoadley C, Hughes JE, Smith P, Allison C, Baron-Cohen S, Simner J (2017) Atypical sensory sensitivity as a shared feature between synaesthesia and autism. Sci Rep 7:41155
Devescovi R, Monasta L, Mancini A, Bin M, Vellante V, Carrozzi M, Colombi C (2016) Early diagnosis and early start denver model intervention in autism spectrum disorders delivered in an italian public health system service. Neuropsychiatr Dis Treat 12:1379–1384
Bhat S, Acharya UR, Adeli H, Bairy GM, Adeli A (2014) Autism: cause factors, early diagnosis and therapies. Rev Neurosci 25:841–850
Georgescu AL, Kuzmanovic B, Roth D, Bente G, Vogeley K (2014) The use of virtual characters to assess and train non-verbal communication in high-functioning autism. Front Hum Neurosci 8:807
Amato CA, Fernandes FD (2010) Interactive use of communication by verbal and non-verbal autistic children. Pro Fono 22:373–378
Vanmarcke S, Mullin C, Van der Hallen R, Evers K, Noens I, Steyaert J, Wagemans J (2016) In the Eye of the beholder: rapid visual perception of real-life scenes by young adults with and without ASD. J Autism Dev Disord 46:2635–2652
Osborne J (2003) Art and the child with autism: therapy or education? Early Child Dev Care 173:411–423
Harris SL (1984) Intervention planning for the family of the autistic child: A multilevel assessment of the family system. J Marital Fam Therapy 10:157–166
Gallagher SA, Gallagher JJ (2002) Giftedness and Asperger’s syndrome: A new agenda for education. Understand Our Gifted 14:7–12
Turner-Brown LM, Lam KS, Holtzclaw TN, Dichter GS, Bodfish JW (2011) Phenomenology and measurement of circumscribed interests in autism spectrum disorders. Autism 15:437–456
Walsh P, Elsabbagh M, Bolton P, Singh I (2011) In search of biomarkers for autism: scientific, social and ethical challenges. Nat Rev Neurosci 12:603–612
Bauman ML (2010) Medical comorbidities in autism: challenges to diagnosis and treatment. Neurotherapeutics 7:320–327
Liu H, Talalay P, Fahey JW (2016) Biomarker-guided strategy for treatment of autism spectrum disorder (ASD). CNS Neurol Disord Drug Targets 15:602–613
Yusuf A, Elsabbagh M (2015) At the cross-roads of participatory research and biomarker discovery in autism: the need for empirical data. BMC Med Ethics 16:88
Sponheim E (1996) Changing criteria of autistic disorders: a comparison of the ICD-10 research criteria and DSM-IV with DSM-III-R, CARS, and ABC. J Autism Dev Disord 26:513–525
Posar A, Visconti P (2017) Autism spectrum disorders: the troubles with the diagnostic and statistical manual of mental disorders 5(th) edition. J Pediatr Neurosci 12:114–115
Foss-Feig JH, Stavropoulos KKM, McPartland JC, Wallace MT, Stone WL, Key AP (2018) Electrophysiological response during auditory gap detection: Biomarker for sensory and communication alterations in autism spectrum disorder? Dev Neuropsychol 43:109–122
Griffin R, Westbury C (2011) Infant EEG activity as a biomarker for autism: a promising approach or a false promise? BMC Med 9:61
Bazelmans T, Jones EJH, Ghods S, Corrigan S, Toth K, Charman T, Webb SJ (2018) Heart rate mean and variability as a biomarker for phenotypic variation in preschoolers with autism spectrum disorder. Autism Res 12(1):39–52
Bertoglio K, Jill James S, Deprey L, Brule N, Hendren RL (2010) Pilot study of the effect of methyl B12 treatment on behavioral and biomarker measures in children with autism. J Altern Complement Med 16:555–560
Hendren RL, James SJ, Widjaja F, Lawton B, Rosenblatt A, Bent S (2016) Randomized, placebo-controlled trial of methyl B12 for children with autism. J Child Adolesc Psychopharmacol 26:774–783
Hu Z, Yang Y, Zhao Y, Yu H, Ying X, Zhou D, Zhong J, Zheng Z, Liu J, Pan R et al (2018) APOE hypermethylation is associated with autism spectrum disorder in a Chinese population. Exp Ther Med 15:4749–4754
Wang Y, Fang Y, Zhang F, Xu M, Zhang J, Yan J, Ju W, Brown WT, Zhong N (2014) Hypermethylation of the enolase gene (ENO2) in autism. Eur J Pediatr 173:1233–1244
Gilani SZ, Tan DW, Russell-Smith SN, Maybery MT, Mian A, Eastwood PR, Shafait F, Goonewardene M, Whitehouse AJ (2015) Sexually dimorphic facial features vary according to level of autistic-like traits in the general population. J Neurodev Disord 7:14
Ren Y, Feng X, Xia X, Zhang Y, Zhang W, Su J, Wang Z, Xu Y, Zhou F (2018) Gender specificity improves the early-stage detection of clear cell renal cell carcinoma based on methylomic biomarkers. Biomark Med 12:607–618
Ren Y, Zhao S, Jiang D, Feng X, Zhang Y, Wei Z, Wang Z, Zhang W, Zhou QF, Li Y et al (2018) Proteomic biomarkers for lung cancer progression. Biomark Med 12:205–215
Xu C, Liu J, Yang W, Shu Y, Wei Z, Zheng W, Feng X, Zhou F (2018) An OMIC biomarker detection algorithm TriVote and its application in methylomic biomarker detection. Epigenomics 10:335–347
Li B, Zhang N, Wang YG, George AW, Reverter A, Li Y (2018) Genomic Prediction of breeding values using a subset of SNPs identified by three machine learning methods. Front Genet 9:237
Ye Y, Zhang R, Zheng W, Liu S, Zhou F (2017) RIFS: a randomly restarted incremental feature selection algorithm. Sci Rep 7:13013
Guyon I, Weston J, Barnhill S, Vapnik V (2002) Gene selection for cancer classification using support vector machines. J Mach Learn 46:389–422
Nikolova O, Moser R, Kemp C, Gonen M, Margolin AA (2017) Modeling gene-wise dependencies improves the identification of drug response biomarkers in cancer studies. Bioinformatics 33:1362–1369
Zhou M, Luo Y, Sun G, Mai G, Zhou F (2015) Constraint programming based biomarker optimization. Biomed Res Int 2015:910515
Liao Z, Wan S, He Y, Zou Q (2018) Classification of small GTPases with hybrid protein features and advanced machine learning techniques. Curr Bioinform 13:492–500
Polewko-Klim A, Lesinski W, Mnich K, Piliszek R, Rudnicki WR (2018) Integration of multiple types of genetic markers for neuroblastoma may contribute to improved prediction of the overall survival. Biol Direct 13:17
Zhang Z, Xu H, Xue Y, Li J, Ye Q (2018) Risk stratification of prostate cancer using the combination of histogram analysis of apparent diffusion coefficient across tumor diffusion volume and clinical information: a pilot study. J Magn Reson Imaging 49(2):556–564
Leger S, Zwanenburg A, Pilz K, Lohaus F, Linge A, Zophel K, Kotzerke J, Schreiber A, Tinhofer I, Budach V et al (2017) A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci Rep 7:13206
Lafzi A, Kazan H (2016) Inferring RBP-mediated regulation in lung squamous cell carcinoma. PLoS One 11:e0155354
Xu X, Zhang X, Tian Q, Wang H, Cui LB, Li S, Tang X, Li B, Dolz J, Ayed IB et al (2018) Quantitative identification of nonmuscle-invasive and muscle-invasive bladder carcinomas: a multiparametric MRI radiomics analysis. J Magn Reson Imaging
Suh HB, Choi YS, Bae S, Ahn SS, Chang JH, Kang SG, Kim EH, Kim SH, Lee SK (2018) Primary central nervous system lymphoma and atypical glioblastoma: Differentiation using radiomics approach. Eur Radiol 28:3832–3839
Alisch RS, Barwick BG, Chopra P, Myrick LK, Satten GA, Conneely KN, Warren ST (2012) Age-associated DNA methylation in pediatric populations. Genome Res 22:623–632
Clough E, Barrett T (2016) The gene expression omnibus database. Methods Mol Biol 1418:93–110
Mokhtari SA, Farzadkia M, Esrafili A, Kalantari RR, Jafari AJ, Kermani M, Gholami M (2016) Bisphenol A removal from aqueous solutions using novel UV/persulfate/H2O2/Cu system: optimization and modelling with central composite design and response surface methodology. J Environ Health Sci Eng 14:19
Mangion K, Gao H, McComb C, Carrick D, Clerfond G, Zhong X, Luo X, Haig CE, Berry C (2016) A novel method for estimating myocardial strain: assessment of deformation tracking against reference magnetic resonance methods in healthy volunteers. Sci Rep 6:38774
Bangdiwala SI (2016) Chi-squared statistics of association and homogeneity. Int J Inj Contr Saf Promot 23:444–446
Wei XX, Stocker AA (2016) Mutual information, fisher information, and efficient coding. Neural Comput 28:305–326
Liu AN, Wang LL, Li HP, Gong J, Liu XH (2017) Correlation between posttraumatic growth and posttraumatic stress disorder symptoms based on pearson correlation coefficient: a meta-analysis. J Nerv Ment Dis 205:380–389
Jankowski KRB, Flannelly KJ, Flannelly LT (2018) The t-test: an influential inferential tool in chaplaincy and other healthcare research. J Health Care Chaplain 24:30–39
Kim TK (2015) T test as a parametric statistic. Korean J Anesthesiol 68:540–546
Shevade SK, Keerthi SS (2003) A simple and efficient algorithm for gene selection using sparse logistic regression. Bioinformatics 19:2246–2253
Boser BE, Guyon IM, Vapnik VN (1992) A training algorithm for optimal margin classifiers. In: Proceedings of the fifth annual workshop on computational learning theory. Pittsburgh, Pennsylvania, USA, July 27–29, pp 144–152
Vapnik VN (1995) Constructing learning algorithms. In: The nature of statistical learning theory, 2nd edn. Springer, New York, pp 119–166
Huang X, Zeng J, Zhou L, Hu C, Yin P, Lin X (2016) A new strategy for analyzing time-series data using dynamic networks: identifying prospective biomarkers of hepatocellular carcinoma. Sci Rep 6:32448
Kim Y, Kim J (2004) Gradient LASSO for feature selection. In: Proceedings of the twenty-first international conference on Machine learning. Banff, Alberta, Canada, July 4–8
Youn E, Jeong MK (2009) Class dependent feature scaling method using naive Bayes classifier for text datamining. Pattern Recogn Lett 30:477–485
Rottmann J, Berbeco R (2014) Using an external surrogate for predictor model training in real-time motion management of lung tumors. Med Phys 41:121706
Barker L, Brown C (2001) Logistic regression when binary predictor variables are highly correlated. Stat Med 20:1431–1442
Cawley GC, Talbot NL (2010) On over-fitting in model selection and subsequent selection bias in performance evaluation. J Mach Learn Rese 11:2079–2107
Inzaule SC, Kityo CM, Siwale M, Akanmu AS, Wellington M, de Jager M, Ive P, Mandaliya K, Stevens W, Boender TS et al (2018) Previous antiretroviral drug use compromises standard first-line HIV therapy and is mediated through drug-resistance. Sci Rep 8:15751
Citak-Er F, Firat Z, Kovanlikaya I, Ture U, Ozturk-Isik E (2018) Machine-learning in grading of gliomas based on multi-parametric magnetic resonance imaging at 3T. Comput Biol Med 99:154–160
Yang CH, Weng ZJ, Chuang LY, Yang CS (2017) Identification of SNP–SNP interaction for chronic dialysis patients. Comput Biol Med 83:94–101
Nejadgholi I, Bolic M (2015) A comparative study of PCA, SIMCA and Cole model for classification of bioimpedance spectroscopy measurements. Comput Biol Med 63:42–51
Rahman MM, Bhuiyan MIH, Hassan AR (2018) Sleep stage classification using single-channel EOG. Comput Biol Med 102:211–220
Olsen RM, Aasvang EK, Meyhoff CS, Dissing Sorensen HB (2018) Towards an automated multimodal clinical decision support system at the post anesthesia care unit. Comput Biol Med 101:15–21
Lu S, Xia Y, Cai W, Fulham M, Feng DD (2017) Alzheimer's Disease Neuroimaging I: Early identification of mild cognitive impairment using incomplete random forest-robust support vector machine and FDG-PET imaging. Comput Med Imaging Graph 60:35–41
Cao J, Wu Z, Ye W, Wang H (2017) Learning functional embedding of genes governed by pair-wised labels. In: 2017 2nd IEEE international conference on computational intelligence and applications (ICCIA), 8–11 Sept. 2017, pp 397–401
Pérez-Díaz N, Ruano-Ordas D, Mendez JR, Galvez JF, Fdez-Riverola F (2012) Rough sets for spam filtering: selecting appropriate decision rules for boundary e-mail classification. Appl Soft Comput 12:3671–3682
Yokoi A, Matsuzaki J, Yamamoto Y, Yoneoka Y, Takahashi K, Shimizu H, Uehara T, Ishikawa M, Ikeda SI, Sonoda T et al (2018) Integrated extracellular microRNA profiling for ovarian cancer screening. Nat Commun 9:4319
Al-Ajlan A, El Allali A (2018) CNN-MGP: convolutional neural networks for metagenomics gene prediction. Interdiscip Sci Comput Life Sci. https://doi.org/10.1007/s12539-018-0313-4
He J, Fang T, Zhang Z, Huang B, Zhu X, Xiong Y (2018) PseUI: pseudouridine sites identification based on RNA sequence information. BMC Bioinform 19:306
Feng X, Zhang R, Liu M, Liu Q, Li F, Yan Z, Zhou F (2019) An accurate regression of developmental stages for breast cancer based on transcriptomic biomarkers. Biomark Med 13:5–15
Xiong Y, Wang Q, Yang J, Zhu X, Wei DQ (2018) PredT4SE-stack: prediction of bacterial type iv secreted effectors from protein sequences using a stacked ensemble method. Front Microbiol 9:2571
Konig C, Alquezar R, Vellido A, Giraldo J (2018) Systematic analysis of primary sequence domain segments for the discrimination between class C GPCR subtypes. Interdiscip Sci 10:43–52
Zhao R, Zhang R, Tang T, Feng X, Li J, Liu Y, Zhu R, Wang G, Li K, Zhou W et al (2018) TriZ-a rotation-tolerant image feature and its application in endoscope-based disease diagnosis. Comput Biol Med 99:182–190
Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, Lally E, Weiss LA, Najm J, Kutsche K et al (2008) Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet 82:199–207
Feng J, Schroer R, Yan J, Song W, Yang C, Bockholt A, Cook EH Jr, Skinner C, Schwartz CE, Sommer SS (2006) High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett 409:10–13
McFarlane HG, Kusek GK, Yang M, Phoenix JL, Bolivar VJ, Crawley JN (2008) Autism-like behavioral phenotypes in BTBR T+tf/J mice. Genes Brain Behav 7:152–163
Raux G, Bumsel E, Hecketsweiler B, van Amelsvoort T, Zinkstok J, Manouvrier-Hanu S, Fantini C, Breviere GM, Di Rosa G, Pustorino G et al (2007) Involvement of hyperprolinemia in cognitive and psychiatric features of the 22q11 deletion syndrome. Hum Mol Genet 16:83–91
Acknowledgements
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13040400), Jilin Provincial Key Laboratory of Big Data Intelligent Computing (20180622002JC), the Education Department of Jilin Province (JJKH20180145KJ), and the start-up grant of the Jilin University. This work was also partially supported by the Bioknow MedAI Institute (BMCPP-2018-001), and the High Performance Computing Center of Jilin University, China. The constructive comments from the two anonymous reviewers were greatly appreciated.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Feng, X., Hao, X., Xin, R. et al. Detecting Methylomic Biomarkers of Pediatric Autism in the Peripheral Blood Leukocytes. Interdiscip Sci Comput Life Sci 11, 237–246 (2019). https://doi.org/10.1007/s12539-019-00328-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-019-00328-9