Abstract
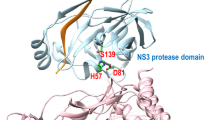
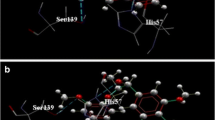
Presently, there are no effective vaccines and anti-virals for the prevention and treatment of Hepatitis C virus infections and hence there is an urgent need to develop potent HCV inhibitors. In this study, we have carried out molecular docking, molecular dynamics and 3D-QSAR on heteroaryl 3-(1,1-dioxo-2H-(1,2,4)-benzothiadizin-3-yl)-4-hydroxy-2(1H)-quinolinone series using NS5B protein. Total of 41 quinolinone derivatives is used for molecular modeling study. The binding conformation and hydrogen bond interaction of the docked complexes were analyzed to model the inhibitors. We identified the molecule XXXV that had a higher affinity with NS5B. The molecular dynamics study confirmed the stability of the compound XXXV-NS5B complex. The developed CoMFA descriptors parameters, which were calculated using a test set of 13 compounds, were statistically significant. Our results will provide useful insights and lead to design potent anti-Hepatitis C virus molecules.






Similar content being viewed by others
Abbreviations
- CoMFA:
-
Comparative Molecular Field Analysis
- HCV:
-
Hepatitis C virus
- NS5B:
-
Nonstructural protein 5B
- ONC:
-
Optimal Number of Components
- PDB:
-
Protein Data Bank
- PLS:
-
Partial Least Square
- QSAR:
-
Quantitative Structure Activity Relationship
- SEE:
-
Standard Error of Estimate
References
Budkowska., A. (2009). Mechanism of cell infection with hepatitis C virus (HCV)-a new paradigm in virus-cell interaction. Polish Journal of Microbiology, 58, 93–98.
Uprichard, S. L. (2010). Hepatitis C virus experimental model systems and antiviral drug research. Virologica Sinica, 25, 227–245.
Chevaliez, S., & Pawlotsky, J. M. (2006). HCV Genome and Life Cycle. In S. L. Tan (Ed.), Hepatitis C Viruses: Genomes and Molecular Biology (pp. 5–47). Norfolk: Horizon Scientific Press.
Luban, N. L., Colvin, C. A., Mohan, P., & Alter, H. J. (2007). The epidemiology of transfusion-associated hepatitis C in a children’s hospital. Transfusion, 47, 615–620.
Nogueira, C. A., Edelman, D. C., Nogueira, C. M., Nogueira, S. A., Coelho, H. S., & Abrahao, L. J., et al. (2002). Hepatitis C virus transfusion-transmitted infection in Brazilian cardiac surgery patients. Clinical Laboratory, 48, 529–533.
Beaulieu, P. L., Bos, M., Bousquet, Y., Fazal, G., Gauthier, J., & Gillard, J., et al. (2004). Non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase: discovery and preliminary SAR of benzimidazole derivatives. Bioorganic & Medicinal Chemistry Letters, 14, 119–124.
Nichols, D. B., Fournet, G., Gurukumar, K. R., Basu, A., Lee, J. C., & Sakamoto, N., et al. (2012). Inhibition of hepatitis C virus NS5B polymerase by S-trityl-L-cysteine derivatives. European Journal of Medicinal Chemistry, 49, 191–199.
Ding, Q., von Schaewen, M., & Ploss, A. (2014). The impact of hepatitis C virus entry on viral tropism. Cell Host & Microbe, 16, 562–568.
Bartenschlager, R., Frese, M., & Pietschmann, T. (2004). Novel insights into hepatitis C virus replication and persistence. Advances in Virus Research, 63, 71–180.
Behrens, S. E., Tomei, L., & De Francesco, R. (1996). Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. The EMBO Journal, 15, 12–22.
Hugle, T., & Cerny, A. (2003). Current therapy and new molecular approaches to antiviral treatment and prevention of hepatitis C. Reviews in Medical Virology, 13, 361–371.
Kang, I. J., Wang, L. W., Hsu, S. J., Lee, C. C., Lee, Y. C., & Wu, Y. S., et al. (2009). Design and efficient synthesis of novel arylthiourea derivatives as potent hepatitis C virus inhibitors. Bioorganic & Medicinal Chemistry Letters, 19, 6063–6068.
Khatri, N., Lather, V., & Madan, A. K. (2015). Diverse classification models for anti-hepatitis C virus activity of thiourea derivatives. Chemometrics and Intelligent Laboratory Systems, 140, 13–21.
Lokwani, D. K., Mokale, S. N., & Shinde, D. B. (2014). 3D QSAR studies based in silico screening of 4,5,6-triphenyl-1,2,3,4-tetrahydropyrimidine analogs for anti-inflammatory activity. European Journal of Medicinal Chemistry, 73, 233–242.
Lavanchy, D. (2012). Viral hepatitis: global goals for vaccination. Journal of Clinical Virology, 55, 296–302.
Aronsohn, A., & Reau, N. (2009). Long-term outcomes after treatment with interferon and ribavirin in HCV patients. Journal of Clinical Gastroenterology, 43, 661–671.
Feuerstadt, P., Bunim, A. L., Garcia, H., Karlitz, J. J., Massoumi, H., & Thosani, A. J., et al. (2010). Effectiveness of hepatitis C treatment with pegylated interferon and ribavirin in urban minority patients. Hepatology, 51, 1137–1143.
Palumbo, E. (2011). Pegylated interferon and ribavirin treatment for hepatitis C virus infection. Therapeutic Advances in Chronic Disease, 2, 39–45.
Chen, J., Zhang, L., Guo, H., Wang, S., Wang, L., Ma, L., & Lu, X. (2014). Activity prediction of hepatitis C virus NS5B polymerase inhibitors of pyridazinone derivatives. Chemometrics and Intelligent Laboratory Systems, 134, 100–109.
Lechmann, M., & Liang, T. J. (2000). Vaccine development for hepatitis C. Seminars in Liver Disease, 20, 211–226.
Beaulieu, P. L. (2009). Recent advances in the development of NS5B polymerase inhibitors for the treatment of hepatitis C virus infection. Expert Opinion on Therapeutic Patents, 19, 145–164.
Delang, L., Froeyen, M., Herdewijn, P., & Neyts, J. (2012). Identification of a novel resistance mutation for benzimidazole inhibitors of the HCV RNA-dependent RNA polymerase. Antiviral Research, 93, 30–38.
Hirashima, S., Suzuki, T., Ishida, T., Noji, S., Yata, S., & Ando, I., et al. (2006). Benzimidazole derivatives bearing substituted biphenyls as hepatitis C virus NS5B RNA-dependent RNA polymerase inhibitors: structure-activity relationship studies and identification of a potent and highly selective inhibitor JTK-109. Journal of Medicinal Chemistry, 49, 4721–4736.
Tomei, L., Altamura, S., Bartholomew, L., Biroccio, A., Ceccacci, A., & Pacini, L., et al. (2003). Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. Journal of Virology, 77, 13225–13231.
Gopalsamy, A., Chopra, R., Lim, K., Ciszewski, G., Shi, M., & Curran, K. J., et al. (2006). Discovery of proline sulfonamides as potent and selective hepatitis C virus NS5b polymerase inhibitors. Evidence for a new NS5b polymerase binding site. Journal of Medicinal Chemistry, 49, 3052–3055.
Rong, F., Chow, S., Yan, S., Larson, G., Hong, Z., & Wu, J. (2007). Structure-activity relationship (SAR) studies of quinoxalines as novel HCV NS5B RNA-dependent RNA polymerase inhibitors. Bioorganic & Medicinal Chemistry Letters, 17, 1663–1666.
Fitch, D. M., Evans, K. A., Chai, D., & Duffy, K. J. (2005). A highly efficient, asymmetric synthesis of benzothiadiazine-substituted tetramic acids: potent inhibitors of hepatitis C virus RNA-dependent RNA polymerase. Organic Letters, 7, 5521–5524.
Harper, S., Avolio, S., Pacini, B., Di Filippo, M., Altamura, S., & Tomei, L., et al. (2005). Potent inhibitors of subgenomic hepatitis C virus RNA replication through optimization of indole-N-acetamide allosteric inhibitors of the viral NS5B polymerase. Journal of Medicinal Chemistry, 48, 4547–4557.
Kumar, K. M., Anbarasu, A., & Ramaiah, S. (2014). Molecular docking and molecular dynamics studies on β-lactamases and penicillin binding proteins. Molecular BioSystems, 10, 891–900.
Parimelzaghan, A., Anbarasu, A., & Ramaiah, S. (2016). Gene network analysis of metallo beta lactamase family proteins indicates the role of gene partners in antibiotic resistance and reveals important drug targets. Journal of Cellular Biochemistry, 117, 1330–1339.
Tedesco, R., Chai, D., Darcy, M. G., Dhanak, D., Fitch, D. M., & Gates, A., et al. (2009). Synthesis and biological activity of heteroaryl 3-(1,1-dioxo-2H-(1,2,4)-benzothiadizin-3-yl)-4-hydroxy-2(1H)-quinolinone derivatives as hepatitis C virus NS5B polymerase inhibitors. Bioorganic & Medicinal Chemistry Letters, 19, 4354–4358.
Tedesco, R., Shaw, A. N., Bambal, R., Chai, D., Concha, N. O., Darcy, M. G., Dhanak, D., Fitch, D. M., Gates, A., Gerhardt, W. G., Halegoua, D. L., Han, C., Hofmann, G. A., Johnston, V. K., Kaura, A. C., Liu, N., Keenan, R. M., Lin-Goerke, J., Sarisky, R. T., Wiggall, K. J., Zimmerman, M. N., & Duffy, K. J. (2006). 3-(1,1-dioxo-2H-(1,2,4)-benzothiadiazin-3-yl)-4-hydroxy-2(1H)-quinolinones, potent inhibitors of hepatitis C virus RNA-dependent RNA polymerase. Journal of Medicinal Chemistry, 49, 971–983.
Li, Z., Wan, H., Shi, Y., & Ouyang, P. (2004). Personal experience with four kinds of chemical structure drawing software: review on ChemDraw, ChemWindow, ISIS/Draw, and ChemSketch. Journal of Chemical Information and Modeling, 44, 1886–1890.
Stammers, T. A., Coulombe, R., Duplessis, M., Fazal, G., Gagnon, A., & Garneau, M., et al. (2013). Anthranilic acid-based Thumb Pocket 2 HCV NS5B polymerase inhibitors with sub-micromolar potency in the cell-based replicon assay. Bioorganic & Medicinal Chemistry Letters, 23, 6879–6885.
Jain, A. N. (2003). Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. Journal of Medicinal Chemistry, 46, 499–511.
Suganya, S., Nandagopal, B., & Anbarasu, A. (2017). Natural inhibitors of HMG-CoA reductase-an in-silico approach through molecular docking and simulation studies. Journal of Cellular Biochemistry, 118, 52–57.
Wei, L., Chintala, S., Ciamporcero, E., Ramakrishnan, S., Elbanna, M., & Wang, J., et al. (2016). Genomic profiling is predictive of response to cisplatin treatment but not to PI3K inhibition in bladder cancer patient-derived xenografts. Oncotarget, 7, 76374–76389.
Malathi, K., & Ramaiah, S. (2016). Molecular Docking and molecular dynamics studies to identify potential OXA-10 Extended Spectrum β-Lactamase non-hydrolysing inhibitors for Pseudomonas aeruginosa. Cell Biochemistry and Biophysics, 74, 141–155.
Gasteiger, J., & Marsili, M. (1980). Iterative partial equalization of orbital electronegativity - a rapid access to atomic charges. Tetrahedron, 36, 3219–3228.
Pearlman, D. A., Case, D. A., Caldwell, J. W., Ross, W. S., Cheatham, III, T. E., & DeBolt, S., et al. (1995). AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Computer Physics Communications, 91, 1–41.
Fletcher, R., & Powell, M. J. D. (1963). A rapidly convergent descent method for minimization. The Computer Journal, 6, 163–168.
Wu, X., Wu, S., & Chen, W. H. (2012). Molecular docking and 3D-QSAR study on 4-(1H-indazol-4-yl) phenylamino and aminopyrazolopyridine urea derivatives as kinase insert domain receptor (KDR) inhibitors. Journal of Molecular Modeling, 18, 1207–1218.
Malathi, K., Anbarasu, A., & Ramaiah, S. (2017). Exploring the resistance mechanism of imipenem in carbapenem hydrolysing class D beta-lactamases OXA-143 and its variant OXA-231 (D224A) expressing Acinetobacter baumannii: An in-silico approach. Computational Biology and Chemistry, 67, 1–8.
Hess, B., Kutzner, C., van der Spoel, D., & Lindahl, E. (2008). GROMACS 4: Algorithms for highly Efficient, Load-Balanced, and Scalable Molecular Simulation. Journal of Chemical Theory and Computation, 4, 435–447.
Oostenbrink, C., Villa, A., Mark, A. E., & van Gunsteren, W. F. (2004). A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. Journal of Computational Chemistry, 25, 1656–1676.
Schuttelkopf, A. W., & van Aalten, D. M. (2004). PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallographica Section D: Biological Crystallography, 60, 1355–1363.
Berendsen, H. J. C., Postama, J. P. M., van Gunsteren, W. F., & Hermans, J. (1981). Interaction models for water in relation to protein hydration. In B. Pullmann (Ed.), Intermolecular Forces (pp. 331–342). Dordrecht: D. Reidel Publishing Company.
Hess, B., Bekker, H., Berendsen, H. J. C., & Fraaije, J. G. E. M. (1997). LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, 18, 1463–1472.
Darden, T., York, D., & Pedersen, L. (1993). Particle mesh Ewald: An Nlog(N) method for Ewald sums in large systems. Journal of Chemical Physics, 98, 10089–10092.
Turner, P. J. (2005). XMGRACE, Version 5.1.19. Beaverton, ORE, USA: Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology.
Thillainayagam, M., Pandian, L., Murugan, K. K., Vijayaparthasarathi, V., Sundaramoorthy, S., Anbarasu, A., & Ramaiah, S. (2015). In silico analysis reveals the anti-malarial potential of quinolinyl chalcone derivatives. Journal of Biomolecular Structure and Dynamics, 33, 961–977.
Thillainayagam, M., Anbarasu, A., & Ramaiah, S. (2016). Comparative molecular field analysis and molecular docking studies on novel aryl chalcone derivatives against an important drug target cysteine protease in Plasmodium falciparum. Journal of Theoretical Biology, 403, 110–128.
Paul, D., Hoppe, S., Saher, G., Krijnse-Locker, J., & Bartenschlager, R. (2013). Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. Journal of Virology, 87, 10612–10627.
Acknowledgements
AA and SR thank Indian Council of Medical Research (ICMR), Government of India Agency for the Research Grant (IRIS ID: 2014-0099) and also thank the management of VIT for the facilities provided.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Malathi, K., Ramaiah, S. & Anbarasu, A. Comparative Molecular Field Analysis and Molecular Docking Studies on Quinolinone Derivatives Indicate Potential Hepatitis C Virus Inhibitors. Cell Biochem Biophys 77, 139–156 (2019). https://doi.org/10.1007/s12013-019-00867-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-019-00867-4