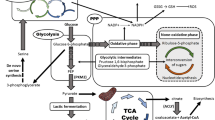
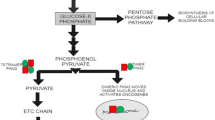
Abstract
Metabolic reprogramming is a newly emerged hallmark of cancer attaining a recent consideration as an essential factor for the progression and endurance of cancer cells. A prime event of this altered metabolism is increased glucose uptake and discharge of lactate into the cells surrounding constructing a favorable tumor niche. Several oncogenic factors help in promoting this consequence including, pyruvate kinase M2 (PKM2) a rate-limiting enzyme of glycolysis in tumor metabolism via exhibiting its low pyruvate kinase activity and nuclear moon-lightening functions to increase the synthesis of lactate and macromolecules for tumor proliferation. Not only its role in cancer cells but also its role in the tumor microenvironment cells has to be understood for developing the small molecules against it which is lacking with the literature till date. Therefore, in this present review, the role of PKM2 with respect to various tumor niche cells will be clarified. Further, it highlights the updated list of therapeutics targeting PKM2 pre-clinically and clinically with their added limitations. This upgraded understanding of PKM2 may provide a pace for the reader in developing chemotherapeutic strategies for better clinical survival with limited resistance.



Similar content being viewed by others
Abbreviations
- ANGPT2:
-
Angiopoietin 2
- AKT:
-
Serine/threonine-specific protein kinase
- ALCL:
-
Anaplastic large cell lymphoma
- ALT:
-
Alanine aminotransferase
- AMPK:
-
5' AMP-activated protein kinase
- ANGPTL4 :
-
Angiopoietin-like 4
- AP-1:
-
Activator protein 1
- C/EBPβ :
-
CCAAT-enhancer binding protein β
- CD4:
-
Cluster of differentiation 4
- COX-2:
-
Cyclooxygenase-2
- CTGF:
-
Connective tissue growth factor
- CXCL8:
-
C-X-C motif chemokine 8
- DASA-58:
-
Dihydro benzodioxin sulfonyl hexahydro diazepin sulfonyl benzenamine
- EGF:
-
Epidermal growth factor
- EMT:
-
Epithelial–mesenchymal transition
- ErbB3:
-
Erb-B2 Receptor tyrosine kinase 3
- ERK-5:
-
Extracellular-signal-regulated kinase 5
- FBP:
-
Fructose bisphosphate
- FGF:
-
Fibroblast growth factor
- FGFR1 :
-
Fibroblast growth factor receptor 1
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- GLUT-1:
-
Glucose transporter -1
- HCT-116:
-
Human colon cancer cell line
- HIF-1α:
-
Hypoxia induced factor-1 alpha
- HK-2:
-
Hexokinase-2
- HMGB1:
-
High mobility group box-1
- IFN-ϒ:
-
Interferon-gamma
- IL-10:
-
Interleukin-10
- IL-13:
-
Interleukin-13
- IL-1β:
-
Interleukin-1β
- IL-4:
-
Interleukin-4
- IL-6:
-
Interleukin-6
- JAK:
-
Janus kinase
- JNK:
-
c-Jun N-terminal kinases
- LDH:
-
Lactate dehydrogenase
- LDH-A:
-
Lactate dehydrogenase-A
- LOX:
-
Lysyl oxidase
- LPS:
-
Lipopolysaccharide
- MAPK:
-
Mitogen-activated protein kinases
- MAP K-5:
-
Mitogen activated protein kinase-5
- MCT-1:
-
Monocarboxylase transporter -1
- MCT4:
-
Monocarboxylate transporter 4
- MDM2:
-
Mouse double minute-2 homolog
- miRNA-124:
-
MicroRNA-124
- MMP:
-
Matrix metalloproteinase
- MMP-2 :
-
Matrix metalloproteinase 2
- mTOR:
-
Mechanistic target of rapamycin
- NF-kB :
-
Nuclear factor kappa B
- NK cells:
-
Natural killer cells
- OXPHOS:
-
Oxidative phosphorylation
- p53:
-
Tumor protein p53
- PD-1:
-
Programmed cell death-1
- PD-4:
-
Programmed cell death-4
- PDGF:
-
Platelet-derived growth factor
- PDGF-BB:
-
Ligand for platelet derived growth factor receptor
- PGAM1:
-
Phosphoglycerate Mutase 1
- PGE2:
-
Prostaglandin E2
- PI3K:
-
Phosphoinositide-3-kinase
- PIN1:
-
Peptidyl-prolyl cis-trans isomerase
- PTP1B:
-
Tyrosine-protein phosphatase non-receptor type 1
- ROS:
-
Reactive oxygen species
- RT-PCR:
-
Real time-polymerase chain reaction
- SAICAR:
-
Succino 5-amino-4-imidazole-N-succinocarboxamide ribonucleotide
- SDF-1:
-
Stromal cell-derived factor 1
- shRNA:
-
Small hairpin ribonucleic acid
- siRNA:
-
Small interfering ribonucleic acid
- SOD2:
-
Superoxide dismutase 2
- STAT3:
-
Signal transducer and activator of transcription 3
- TCA:
-
Tricarboxylic acid
- TCR:
-
T cell receptor
- TEPP-46:
-
Thieno-pyrrole-pyridazine
- TGFα:
-
Transforming growth factor alpha
- TGFβ:
-
Transforming growth factor beta
- TH-1:
-
T helper cell-1
- TIMP:
-
Tissue inhibitor of metalloproteinases
- TNF-α:
-
Tumor necrosis factor-alpha
- TRAF-5:
-
TNF-receptor associated factor-5
- US28:
-
G-protein coupled receptor homolog US28
- VEGF:
-
Vascular endothelial growth factor
- vGPCR :
-
Viral G protein-coupled receptor
References
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA: A Cancer Journal for Clinicians 68 (1):7-30. https://doi.org/10.3322/caac.21442
Rübben A, Araujo A (2017) Cancer heterogeneity: converting a limitation into a source of biologic information. J Transl Med 15(1):190
Brown TM, Fee E (2006) Rudolf Carl Virchow: medical scientist, social reformer, role model. Am J Public Health 96(12):2104–2105
Seth S, Li C-Y, Ho I-L, Corti D, Loponte S, Sapio L, Del Poggetto E, Yen E-Y, Robinson FS, Peoples M (2019) Pre-existing Functional Heterogeneity of Tumorigenic Compartment as the Origin of Chemoresistance in Pancreatic Tumors. Cell Rep 26(6):1518–1532. e1519
Kang J, Chen HJ, Zhang XC, Su J, Zhou Q, Tu HY, Wang Z, Wang BC, Zhong WZ, Yang XN (2018) Heterogeneous responses and resistant mechanisms to crizotinib in ALK-positive advanced non-small cell lung cancer. Thorac Cancer 9(9):1093–1103
Nalla LV, Kalia K, Khairnar A (2019) Self-renewal signaling pathways in breast cancer stem cells. Int J Biochem Cell Biol 107:140–153
Marusyk A, Polyak K (2010) Tumor heterogeneity: causes and consequences. Biochim Biophys Acta 1805(1):105–117. https://doi.org/10.1016/j.bbcan.2009.11.002
Fisher R, Pusztai L, Swanton C (2013) Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer 108:479. https://doi.org/10.1038/bjc.2012.581
Dagogo-Jack I, Shaw AT (2017) Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 15:81. https://doi.org/10.1038/nrclinonc.2017.166
Neugent ML, Goodwin J, Sankaranarayanan I, Yetkin CE, Hsieh M-H, J-w K (2018) A new perspective on the heterogeneity of cancer glycolysis. Biomol Ther 26(1):10
Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, Paulson TG, Blount PL, Risques RA, Rabinovitch PS, Reid BJ (2006) Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet 38(4):468–473. https://doi.org/10.1038/ng1768
Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M (2011) Tumour evolution inferred by single-cell sequencing. Nature 472(7341):90–94. https://doi.org/10.1038/nature09807
Bochtler T, Stolzel F, Heilig CE, Kunz C, Mohr B, Jauch A, Janssen JW, Kramer M, Benner A, Bornhauser M, Ho AD, Ehninger G, Schaich M, Kramer A (2013) Clonal heterogeneity as detected by metaphase karyotyping is an indicator of poor prognosis in acute myeloid leukemia. J Clin Oncol Off J Am Soc Clin Oncol 31(31):3898–3905. https://doi.org/10.1200/jco.2013.50.7921
Almendro V, Marusyk A, Polyak K (2013) Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol 8:277–302. https://doi.org/10.1146/annurev-pathol-020712-163923
Bidkhori G, Benfeitas R, Klevstig M, Zhang C, Nielsen J, Uhlen M, Boren J, Mardinoglu A (2018) Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes. Proc Natl Acad Sci 115(50):E11874–E11883
Lamb R, Ozsvari B, Bonuccelli G, Smith DL, Pestell RG, Martinez-Outschoorn UE, Clarke RB, Sotgia F, Lisanti MP (2015) Dissecting tumor metabolic heterogeneity: Telomerase and large cell size metabolically define a sub-population of stem-like, mitochondrial-rich, cancer cells. Oncotarget 6(26):21892
Robertson-Tessi M, Gillies RJ, Gatenby RA, Anderson AR (2015) Impact of metabolic heterogeneity on tumor growth, invasion, and treatment outcomes. Cancer Res 75(8):1567–1579
Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L (2016) Metabolic heterogeneity in human lung tumors. Cell 164(4):681–694
Yuan Y (2016) Spatial heterogeneity in the tumor microenvironment. Cold Spring Harb Perspect Med 6(8):a026583
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674
Chen G, Zhang Y, Liang J, Li W, Zhu Y, Zhang M, Wang C, Hou J (2018) Deregulation of Hexokinase II Is Associated with Glycolysis, Autophagy, and the Epithelial-Mesenchymal Transition in Tongue Squamous Cell Carcinoma under Hypoxia. BioMed research international 2018
Kachel P, Trojanowicz B, Sekulla C, Prenzel H, Dralle H, Hoang-Vu C (2015) Phosphorylation of pyruvate kinase M2 and lactate dehydrogenase A by fibroblast growth factor receptor 1 in benign and malignant thyroid tissue. BMC Cancer 15(1):140
Mogi A, Koga K, Aoki M, Hamasaki M, Uesugi N, Iwasaki A, Shirakusa T, Tamura K, Nabeshima K (2013) Expression and role of GLUT-1, MCT-1, and MCT-4 in malignant pleural mesothelioma. Virchows Archiv 462(1):83–93
Miranda-Gonçalves V, Granja S, Martinho O, Honavar M, Pojo M, Costa BM, Pires MM, Pinheiro C, Cordeiro M, Bebiano G (2016) Hypoxia-mediated upregulation of MCT1 expression supports the glycolytic phenotype of glioblastomas. Oncotarget 7(29):46335
Zha X, Sun Q, Zhang H (2011) mTOR upregulation of glycolytic enzymes promotes tumor development. Taylor & Francis
Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW (2013) PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell 155(2):397–409
Sounni NE, Noel A (2013) Targeting the tumor microenvironment for cancer therapy. Clin Chem 59(1):85–93
Yang L, Lin PC (2017) Mechanisms that drive inflammatory tumor microenvironment, tumor heterogeneity, and metastatic progression. In: Seminars in cancer biology. Elsevier, pp 185–195
Schowen RL (1993) Principles of biochemistry 2nd ed. (Lehninger, Albert L.; Nelson, David L.; Cox, Michael M.). Journal of Chemical Education 70 (8):A223. https://doi.org/10.1021/ed070pA223.1
Locasale JW, Cantley LC (2010) Altered metabolism in cancer. BMC Biology 8(1):88. https://doi.org/10.1186/1741-7007-8-88
Phan LM, Yeung S-CJ, Lee M-H (2014) Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol Med 11(1):1–19. https://doi.org/10.7497/j.issn.2095-3941.2014.01.001
Warburg O (1925) The Metabolism of Carcinoma Cells. J Cancer Res 9(1):148–163. https://doi.org/10.1158/jcr.1925.148
Hu Y, Lu W, Chen G, Wang P, Chen Z, Zhou Y, Ogasawara M, Trachootham D, Feng L, Pelicano H, Chiao PJ, Keating MJ, Garcia-Manero G, Huang P (2012) K-ras(G12V) transformation leads to mitochondrial dysfunction and a metabolic switch from oxidative phosphorylation to glycolysis. Cell Res 22(2):399–412. https://doi.org/10.1038/cr.2011.145
Suzuki S, Okada M, Takeda H, Kuramoto K, Sanomachi T, Togashi K, Seino S, Yamamoto M, Yoshioka T, Kitanaka C (2018) Involvement of GLUT1-mediated glucose transport and metabolism in gefitinib resistance of non-small-cell lung cancer cells. Oncotarget 9(66):32667–32679. https://doi.org/10.18632/oncotarget.25994
Šmerc A, Sodja E, Legiša M (2011) Posttranslational modification of 6-phosphofructo-1-kinase as an important feature of cancer metabolism. PloS one 6(5):e19645
Wiese EK, Hitosugi T (2018) Tyrosine kinase signaling in cancer metabolism: PKM2 paradox in the warburg effect. Front Cell Dev Biol 6
Hallows WC, Yu W, Denu JM (2012) Regulation of glycolytic enzyme phosphoglycerate mutase-1 by Sirt1 protein-mediated deacetylation. J Biol Chem 287(6):3850–3858
Ganapathy-Kanniappan S, Geschwind JF (2013) Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer 12:152. https://doi.org/10.1186/1476-4598-12-152
Li H, Xu H, Xing R, Pan Y, Li W, Cui J, Lu Y (2019) Pyruvate kinase M2 contributes to cell growth in gastric cancer via aerobic glycolysis. Pathology-Research and Practice
Boukouris AE, Zervopoulos SD, Michelakis ED (2016) Metabolic enzymes moonlighting in the nucleus: metabolic regulation of gene transcription. Trends Biochem Sci 41(8):712–730
Dayton TL, Jacks T, Vander Heiden MG (2016) PKM2, cancer metabolism, and the road ahead. EMBO Rep 17(12):1721–1730
Mazurek S, Boschek CB, Hugo F, Eigenbrodt E (2005) Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol 15(4):300–308. https://doi.org/10.1016/j.semcancer.2005.04.009
Altenberg B, Greulich KO (2004) Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 84(6):1014–1020. https://doi.org/10.1016/j.ygeno.2004.08.010
Iqbal MA, Gupta V, Gopinath P, Mazurek S, Bamezai RN (2014) Pyruvate kinase M2 and cancer: an updated assessment. FEBS Letters 588(16):2685–2692. https://doi.org/10.1016/j.febslet.2014.04.011
David CJ, Chen M, Assanah M, Canoll P, Manley JL (2010) HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463(7279):364
Semenza G (2011) Regulation of metabolism by hypoxia-inducible factor 1. In: Cold Spring Harbor symposia on quantitative biology. Cold Spring Harbor Laboratory Press, pp 347–353
Dombrauckas JD, Santarsiero BD, Mesecar AD (2005) Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry 44(27):9417–9429
Mazurek S (2011) Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol 43(7):969–980
Hitosugi T, Kang S, Vander Heiden MG, Chung T-W, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ (2009) Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal 2(97):ra73-ra73
Yang W, Lu Z (2013) Regulation and function of pyruvate kinase M2 in cancer. Cancer Lett 339(2):153–158. https://doi.org/10.1016/j.canlet.2013.06.008
Warner SL, Carpenter KJ, Bearss DJ (2014) Activators of PKM2 in cancer metabolism. Future Med Chem 6(10):1167–1178. https://doi.org/10.4155/fmc.14.70
Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, Lu Z (2012) ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol 14(12):1295
Gao X, Wang H, Yang JJ, Liu X, Liu ZR (2012) Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell 45(5):598–609. https://doi.org/10.1016/j.molcel.2012.01.001
Yang P, Li Z, Fu R, Wu H, Li Z (2014) Pyruvate kinase M2 facilitates colon cancer cell migration via the modulation of STAT3 signalling. Cell Signal 26(9):1853–1862. https://doi.org/10.1016/j.cellsig.2014.03.020
Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z (2012) PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 150(4):685–696. https://doi.org/10.1016/j.cell.2012.07.018
Wang H-J, Hsieh Y-J, Cheng W-C, Lin C-P, Lin Y-s, Yang S-F, Chen C-C, Izumiya Y, Yu J-S, Kung H-J (2014) JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1α–mediated glucose metabolism. Proc Natl Acad Sci 111 (1):279-284
Li Q, Cao L, Tian Y, Zhang P, Ding C, Lu W, Jia C, Shao C, Liu W, Wang D, Ye H, Hao H (2018) Butyrate Suppresses the Proliferation of Colorectal Cancer Cells via Targeting Pyruvate Kinase M2 and Metabolic Reprogramming. Mol Cell Proteomics 17(8):1531–1545. https://doi.org/10.1074/mcp.RA118.000752
Chen J, Xie J, Jiang Z, Wang B, Wang Y, Hu X (2011) Shikonin and its analogs inhibit cancer cell glycolysis by targeting tumor pyruvate kinase-M2. Oncogene 30(42):4297
Babu MS, Mahanta S, Lakhter AJ, Hato T, Paul S, Naidu SR (2018) Lapachol inhibits glycolysis in cancer cells by targeting pyruvate kinase M2. PloS one 13(2):e0191419
Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, Yang H, Mattaini KR, Metallo CM, Fiske BP, Courtney KD, Malstrom S, Khan TM, Kung C, Skoumbourdis AP, Veith H, Southall N, Walsh MJ, Brimacombe KR, Leister W, Lunt SY, Johnson ZR, Yen KE, Kunii K, Davidson SM, Christofk HR, Austin CP, Inglese J, Harris MH, Asara JM, Stephanopoulos G, Salituro FG, Jin S, Dang L, Auld DS, Park HW, Cantley LC, Thomas CJ, Vander Heiden MG (2012) Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol 8(10):839–847. https://doi.org/10.1038/nchembio.1060
DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB (2008) The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7(1):11–20. https://doi.org/10.1016/j.cmet.2007.10.002
Sun L, Suo C, Li S-t, Zhang H, Gao P (2018) Metabolic reprogramming for cancer cells and their microenvironment: Beyond the Warburg Effect. Biochim Biophys Acta 1870(1):51–66. https://doi.org/10.1016/j.bbcan.2018.06.005
Tennant DA, Durán RV, Gottlieb E (2010) Targeting metabolic transformation for cancer therapy. Nat Rev Cancer 10:267. https://doi.org/10.1038/nrc2817
Li K, Huang S-h, X-m L, Yang L, G-q L, Y-j L (2018) Interaction of cancer cell-derived Foxp3 and tumor microenvironment in human tongue squamous cell carcinoma. Exp Cell Res 370(2):643–652. https://doi.org/10.1016/j.yexcr.2018.07.029
Meurette O, Mehlen P (2018) Notch Signaling in the Tumor Microenvironment. Cancer Cell. https://doi.org/10.1016/j.ccell.2018.07.009
Echizen K, Oshima H, Nakayama M, Oshima M (2018) The inflammatory microenvironment that promotes gastrointestinal cancer development and invasion. Adv Biol Regul 68:39–45. https://doi.org/10.1016/j.jbior.2018.02.001
Ding Z-Y, Zou X-L, Wei Y-Q (2012) Cancer Microenvironment and Cancer Vaccine. Cancer Microenviron 5(3):333–344. https://doi.org/10.1007/s12307-012-0107-x
Taube JM, Klein AP, Brahmer JR, Xu H, Pan X, Kim JH, Chen L, Pardoll DM, Topalian SL, Anders RA (2014) Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res
Ivey JW, Bonakdar M, Kanitkar A, Davalos RV, Verbridge SS (2016) Improving cancer therapies by targeting the physical and chemical hallmarks of the tumor microenvironment. Cancer Lett 380(1):330–339. https://doi.org/10.1016/j.canlet.2015.12.019
Koontongkaew S (2013) The tumor microenvironment contribution to development, growth, invasion and metastasis of head and neck squamous cell carcinomas. J Cancer 4(1):66–83. https://doi.org/10.7150/jca.5112
Subramaniam R, Mizoguchi A, Mizoguchi E (2016) Mechanistic roles of epithelial and immune cell signaling during the development of colitis-associated cancer. Cancer Res Front 2(1):1–21. https://doi.org/10.17980/2016.1
Buetti-Dinh A, O’Hare T, Friedman R (2016) Sensitivity Analysis of the NPM-ALK Signalling Network Reveals Important Pathways for Anaplastic Large Cell Lymphoma Combination Therapy. PLoS ONE 11(9):e0163011. https://doi.org/10.1371/journal.pone.0163011
Liang J, Cao R, Wang X, Zhang Y, Wang P, Gao H, Li C, Yang F, Zeng R, Wei P (2017) Mitochondrial PKM2 regulates oxidative stress-induced apoptosis by stabilizing Bcl2. Cell Res 27(3):329
Li Y-H, Li X-F, Liu J-T, Wang H, Fan L-L, Li J, Sun G-P (2018) PKM2, a potential target for regulating cancer. Gene 668:48–53
Wong N, Ojo D, Yan J, Tang D (2015) PKM2 contributes to cancer metabolism. Cancer Lett 356(2):184–191
Dayton TL, Jacks T, Vander Heiden MG (2016) PKM2, cancer metabolism, and the road ahead. EMBO Rep 17:1721–1730
Wang R, Green DR (2012) The immune diet: meeting the metabolic demands of lymphocyte activation. F1000 Biol Rep 4:9. https://doi.org/10.3410/b4-9
Finlay DK (2013) mTORC1 regulates CD8+ T-cell glucose metabolism and function independently of PI3K and PKB. Biochem Soc Trans 41(2):681–686. https://doi.org/10.1042/bst20120359
MacIver NJ, Michalek RD, Rathmell JC (2013) Metabolic regulation of T lymphocytes. Annu Rev Immunol 31:259–283
Gong J, Chehrazi-Raffle A, Reddi S, Salgia R (2018) Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. J Immunother Cancer 6(1):8. https://doi.org/10.1186/s40425-018-0316-z
Palsson-McDermott EM, Dyck L, Zaslona Z, Menon D, McGettrick AF, Mills KHG, O'Neill LA (2017) Pyruvate Kinase M2 Is Required for the Expression of the Immune Checkpoint PD-L1 in Immune Cells and Tumors. Front Immunol 8:1300. https://doi.org/10.3389/fimmu.2017.01300
Yang W, Lu Z (2013) Nuclear PKM2 regulates the Warburg effect. Cell Cycle (Georgetown, Tex) 12(19):3154–3158. https://doi.org/10.4161/cc.26182
Filipp FV (2013) Cancer metabolism meets systems biology: Pyruvate kinase isoform PKM2 is a metabolic master regulator. J Carcinog 12:14. https://doi.org/10.4103/1477-3163.115423
Luan W, Wang Y, Chen X, Shi Y, Wang J, Zhang J, Qian J, Li R, Tao T, Wei W, Hu Q, Liu N, You Y (2015) PKM2 promotes glucose metabolism and cell growth in gliomas through a mechanism involving a let-7a/c-Myc/hnRNPA1 feedback loop. Oncotarget 6(15):13006–13018. https://doi.org/10.18632/oncotarget.3514
Kimura A, Kitamura K, Ailiken G, Satoh M, Minamoto T, Tanaka N, Nomura F, Matsushita K (2017) FIR haplodeficiency promotes splicing to pyruvate kinase M2 in mice thymic lymphoma tissues revealed by six-plex tandem mass tag quantitative proteomic analysis. Oncotarget 8(40):67955
Batdorf B, Kroft S, Hosking P, Harrington A, Mackinnon A, Olteanu H (2014) Evaluation of CD43 Expression in Non-Hematologic Malignancies. Am J Clin Pathol 142(suppl_1):A244–A244. https://doi.org/10.1093/ajcp/142.suppl1.244
Bravo-Adame ME, Vera-Estrella R, Barkla BJ (2017) An alternative mode of CD43 signal transduction activates pro-survival pathways of T lymphocytes. 150 (1):87-99. https://doi.org/10.1111/imm.12670
Fu Q, Cash S, Andersen J, Kennedy C, Madadi A, Raghavendra M, Dietrich L, Agger W, Shelley C (2014) Intracellular patterns of sialophorin expression define a new molecular classification of breast cancer and represent new targets for therapy. British J Cancer 110(1):146
Fu Q, Cash SE, Andersen JJ, Kennedy CR, Oldenburg DG, Zander VB, Foley GR, Simon Shelley C (2013) CD43 in the nucleus and cytoplasm of lung cancer is a potential therapeutic target. Int J Cancer 132(8):1761–1770
Ranger AM, Gerstenfeld LC, Wang J, Kon T, Bae H, Gravallese EM, Glimcher MJ, Glimcher LH (2000) The nuclear factor of activated T cells (NFAT) transcription factor NFATp (NFATc2) is a repressor of chondrogenesis. J Exp Med 191(1):9–22
Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, Kleinstein SH, Abel ED, Insogna KL, Feske S, Locasale JW, Bosenberg MW, Rathmell JC, Kaech SM (2015) Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell 162(6):1217–1228. https://doi.org/10.1016/j.cell.2015.08.012
Qin J-J, Wang W, Voruganti S, Wang H, Zhang W-D, Zhang R (2015) Inhibiting NFAT1 for breast cancer therapy: new insights into the mechanism of action of MDM2 inhibitor JapA. Oncotarget 6(32):33106
Hwang SR, Murga-Zamalloa C, Brown N, Basappa J, McDonnell SR, Mendoza-Reinoso V, Basrur V, Wilcox R, Elenitoba-Johnson K, Lim MS (2017) Pyrimidine tract-binding protein 1 mediates pyruvate kinase M2-dependent phosphorylation of signal transducer and activator of transcription 3 and oncogenesis in anaplastic large cell lymphoma. Lab Investig 97(8):962
Lu S, Deng J, Liu H, Liu B, Yang J, Miao Y, Li J, Wang N, Jiang C, Xu Q, Wang X, Feng J (2018) PKM2-dependent metabolic reprogramming in CD4(+) T cells is crucial for hyperhomocysteinemia-accelerated atherosclerosis. J Mol Med (Berlin, Germany) 96(6):585–600. https://doi.org/10.1007/s00109-018-1645-6
Lin J, Lee I-M, Song Y, Cook NR, Selhub J, Manson JE, Buring JE, Zhang SM (2010) Plasma Homocysteine and Cysteine and Risk of Breast Cancer in Women. Cancer Res 70(6):2397–2405. https://doi.org/10.1158/0008-5472.can-09-3648
Hoffman W, Lakkis FG, Chalasani G (2016) B Cells, Antibodies, and More. Clin J Am Soc Nephrol: CJASN 11(1):137–154. https://doi.org/10.2215/CJN.09430915
Sugio T, Miyawaki K, Kato K, Sasaki K, Yamada K, Iqbal J, Miyamoto T, Ohshima K, Maeda T, Miyoshi H, Akashi K (2018) Microenvironmental immune cell signatures dictate clinical outcomes for PTCL-NOS. Blood Adv 2(17):2242–2252. https://doi.org/10.1182/bloodadvances.2018018754
Deng J, Lü S, Liu H, Liu B, Jiang C, Xu Q, Feng J, Wang X (2017) Homocysteine Activates B Cells via Regulating PKM2-Dependent Metabolic Reprogramming. J Immunol Author Choice 198(1):170–183. https://doi.org/10.4049/jimmunol.1600613
Murray PJ, Wynn TA (2011) Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol 11(11):723–737. https://doi.org/10.1038/nri3073
Martinez FO, Gordon S (2014) The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 6:13. https://doi.org/10.12703/p6-13
Mosser DM, Edwards JP (2008) Exploring the full spectrum of macrophage activation. Nat Rev Immunol 8(12):958–969. https://doi.org/10.1038/nri2448
Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MA, Sheedy FJ, Gleeson LE, van den Bosch MW, Quinn SR, Domingo-Fernandez R, Johnston DG, Jiang JK, Israelsen WJ, Keane J, Thomas C, Clish C, Vander Heiden M, Xavier RJ, O'Neill LA (2015) Pyruvate kinase M2 regulates Hif-1alpha activity and IL-1beta induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab 21(1):65–80. https://doi.org/10.1016/j.cmet.2014.12.005
Corcoran SE, O'Neill LA (2016) HIF1alpha and metabolic reprogramming in inflammation. J Clin Invest 126(10):3699–3707. https://doi.org/10.1172/jci84431
Liou G-Y, Storz P (2010) Reactive oxygen species in cancer. Free Radic Res 44(5):479–496. https://doi.org/10.3109/10715761003667554
Shirai T, Nazarewicz RR, Wallis BB, Yanes RE, Watanabe R, Hilhorst M, Tian L, Harrison DG, Giacomini JC, Assimes TL, Goronzy JJ, Weyand CM (2016) The glycolytic enzyme PKM2 bridges metabolic and inflammatory dysfunction in coronary artery disease. J Exp Med 213(3):337–354. https://doi.org/10.1084/jem.20150900
Li W, Saud SM, Young MR, Chen G, Hua B (2015) Targeting AMPK for cancer prevention and treatment. Oncotarget 6(10):7365–7378. https://doi.org/10.18632/oncotarget.3629
Huang J, Liu K, Zhu S, Xie M, Kang R, Cao L, Tang D (2018) AMPK regulates immunometabolism in sepsis. Brain Behav Immun 72:89–100. https://doi.org/10.1016/j.bbi.2017.11.003
Zhang Z, Deng W, Kang R, Xie M, Billiar T, Wang H, Cao L, Tang D (2016) Plumbagin Protects Mice from Lethal Sepsis by Modulating Immunometabolism Upstream of PKM2. Molecular medicine. Cambridge, Mass. https://doi.org/10.2119/molmed.2015.00250
Lu W, Zuo Y, Feng Y, Zhang M (2014) SIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumour Biol 35(11):10699–10705. https://doi.org/10.1007/s13277-014-2372-4
Wang F, Wang K, Xu W, Zhao S, Ye D, Wang Y, Xu Y, Zhou L, Chu Y, Zhang C, Qin X, Yang P, Yu H (2017) SIRT5 Desuccinylates and Activates Pyruvate Kinase M2 to Block Macrophage IL-1beta Production and to Prevent DSS-Induced Colitis in Mice. Cell Rep 19(11):2331–2344. https://doi.org/10.1016/j.celrep.2017.05.065
Cheng Y, Feng Y, Xia Z, Li X, Rong J (2017) omega-Alkynyl arachidonic acid promotes anti-inflammatory macrophage M2 polarization against acute myocardial infarction via regulating the cross-talk between PKM2, HIF-1alpha and iNOS. Biochim Biophys Acta 1862(12):1595–1605. https://doi.org/10.1016/j.bbalip.2017.09.009
Collin M, McGovern N, Haniffa M (2013) Human dendritic cell subsets. Immunology 140(1):22–30. https://doi.org/10.1111/imm.12117
Gabrilovich DI, Ostrand-Rosenberg S, Bronte V (2012) Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol 12(4):253–268. https://doi.org/10.1038/nri3175
Lee JA, Sung HN, Jeon CH, Gill BC, Oh GS, Youn HJ, Park JH (2008) AIP1, a carbohydrate fraction from Artemisia iwayomogi, modulates the functional differentiation of bone marrow-derived dendritic cells. Int Immunopharmacol 8(4):534–541. https://doi.org/10.1016/j.intimp.2007.12.005
Vannini F, Kashfi K, Nath N (2015) The dual role of iNOS in cancer. Redox Biol 6:334–343. https://doi.org/10.1016/j.redox.2015.08.009
Jiang H, Zhang S, Song T, Guan X, Zhang R, Chen X (2018) Trichostatin a Protects Dendritic Cells Against Oxygen-Glucose Deprivation via the SRSF3/PKM2/Glycolytic Pathway. Front Pharmacol 9:612. https://doi.org/10.3389/fphar.2018.00612
Bronte V, Serafini P, Mazzoni A, Segal DM, Zanovello P (2003) L-arginine metabolism in myeloid cells controls T-lymphocyte functions. Trends Immunol 24(6):301–305. https://doi.org/10.1016/S1471-4906(03)00132-7
Liu WR, Tian MX, Yang LX, Lin YL, Jin L, Ding ZB, Shen YH, Peng YF, Gao DM, Zhou J, Qiu SJ, Dai Z, He R, Fan J, Shi YH (2015) PKM2 promotes metastasis by recruiting myeloid-derived suppressor cells and indicates poor prognosis for hepatocellular carcinoma. Oncotarget 6(2):846–861. https://doi.org/10.18632/oncotarget.2749
Trautwein C, Rakemann T, Malek NP, Plümpe J, Tiegs G, Manns MP (1998) Concanavalin A-induced liver injury triggers hepatocyte proliferation. J Clin Invest 101(9):1960–1969. https://doi.org/10.1172/JCI504
Weltzsch JP, Krech T, Vander Heiden MG, Tiegs G, Horst A (2018) Activation of pyruvate kinase isoform M2 (PKM2) in myeloid cells protects from Concanavalin A-mediated liver injury. J Hepatol 68:S448. https://doi.org/10.1016/S0168-8278(18)31136-X
Paul S, Lal G (2017) The Molecular Mechanism of Natural Killer Cells Function and Its Importance in Cancer Immunotherapy. Front Immunol 8:1124. https://doi.org/10.3389/fimmu.2017.01124
Shi F-D, Ljunggren H-G, La Cava A, Van Kaer L (2011) Organ-specific features of natural killer cells. Nat Rev Immunol 11(10):658–671. https://doi.org/10.1038/nri3065
Tu MM, Mahmoud AB, Makrigiannis AP (2016) Licensed and Unlicensed NK Cells: Differential Roles in Cancer and Viral Control. Front Immunol 7:166–166. https://doi.org/10.3389/fimmu.2016.00166
Schafer JR, Salzillo TC, Chakravarti N, Kararoudi MN, Trikha P, Foltz JA, Wang R, Li S, Lee DA (2019) Education-dependent activation of glycolysis promotes the cytolytic potency of licensed human natural killer cells. J Allergy Clin Immunol 143:346–358. e6
Tang D, Kang R, Zeh HJ 3rd, Lotze MT (2010) High-mobility group box 1 and cancer. Biochim Biophys Acta 1799(1-2):131–140. https://doi.org/10.1016/j.bbagrm.2009.11.014
Gdynia G, Sauer SW, Kopitz J, Fuchs D, Duglova K, Ruppert T, Miller M, Pahl J, Cerwenka A, Enders M, Mairbaurl H, Kaminski MM, Penzel R, Zhang C, Fuller JC, Wade RC, Benner A, Chang-Claude J, Brenner H, Hoffmeister M, Zentgraf H, Schirmacher P, Roth W (2016) The HMGB1 protein induces a metabolic type of tumour cell death by blocking aerobic respiration. Nat Commun 7:10764. https://doi.org/10.1038/ncomms10764
Tao L, Huang G, Song H, Chen Y, Chen L (2017) Cancer associated fibroblasts: An essential role in the tumor microenvironment. Oncol Lett 14(3):2611–2620. https://doi.org/10.3892/ol.2017.6497
LeBleu VS, Kalluri R (2018) A peek into cancer-associated fibroblasts: origins, functions and translational impact. Dis Model Mech 11(4):dmm029447. https://doi.org/10.1242/dmm.029447
Martinez-Outschoorn UE, Lisanti MP, Sotgia F (2014) Catabolic cancer-associated fibroblasts transfer energy and biomass to anabolic cancer cells, fueling tumor growth. Semin Cancer Biol 25:47–60. https://doi.org/10.1016/j.semcancer.2014.01.005
Huang L, Xu AM, Liu S, Liu W, Li T-J (2014) Cancer-associated fibroblasts in digestive tumors. World J Gastroenterol: WJG 20(47):17804–17818. https://doi.org/10.3748/wjg.v20.i47.17804
Radisky DC, Kenny PA, Bissell MJ (2007) Fibrosis and cancer: do myofibroblasts come also from epithelial cells via EMT? J Cell Biochem 101(4):830–839. https://doi.org/10.1002/jcb.21186
Ishikawa M, Inoue T, Shirai T, Takamatsu K, Kunihiro S, Ishii H, Nishikata T (2014) Simultaneous expression of cancer stem cell-like properties and cancer-associated fibroblast-like properties in a primary culture of breast cancer cells. Cancers 6(3):1570–1578. https://doi.org/10.3390/cancers6031570
de Wit RH, Mujic-Delic A, van Senten JR, Fraile-Ramos A, Siderius M, Smit MJ (2016) Human cytomegalovirus encoded chemokine receptor US28 activates the HIF-1alpha/PKM2 axis in glioblastoma cells. Oncotarget 7(42):67966–67985. https://doi.org/10.18632/oncotarget.11817
Kachel P, Trojanowicz B, Sekulla C, Prenzel H, Dralle H, Hoang-Vu C (2015) Phosphorylation of pyruvate kinase M2 and lactate dehydrogenase A by fibroblast growth factor receptor 1 in benign and malignant thyroid tissue. BMC Cancer 15:140. https://doi.org/10.1186/s12885-015-1135-y
Chiavarina B, Whitaker-Menezes D, Martinez-Outschoorn UE, Witkiewicz AK, Birbe R, Howell A, Pestell RG, Smith J, Daniel R, Sotgia F, Lisanti MP (2011) Pyruvate kinase expression (PKM1 and PKM2) in cancer-associated fibroblasts drives stromal nutrient production and tumor growth. Cancer Biol Ther 12(12):1101–1113. https://doi.org/10.4161/cbt.12.12.18703
Chatterjee M, Ben-Josef E, Thomas DG, Morgan MA, Zalupski MM, Khan G, Andrew Robinson C, Griffith KA, Chen C-S, Ludwig T, Bekaii-Saab T, Chakravarti A, Williams TM (2015) Caveolin-1 is Associated with Tumor Progression and Confers a Multi-Modality Resistance Phenotype in Pancreatic Cancer. Sci Rep 5:10867–10867. https://doi.org/10.1038/srep10867
Martinez-Outschoorn UE, Balliet RM, Rivadeneira DB, Chiavarina B, Pavlides S, Wang C, Whitaker-Menezes D, Daumer KM, Lin Z, Witkiewicz AK, Flomenberg N, Howell A, Pestell RG, Knudsen ES, Sotgia F, Lisanti MP (2010) Oxidative stress in cancer associated fibroblasts drives tumor-stroma co-evolution: A new paradigm for understanding tumor metabolism, the field effect and genomic instability in cancer cells. Cell Cycle (Georgetown, Tex) 9(16):3256–3276. https://doi.org/10.4161/cc.9.16.12553
Santi A, Caselli A, Ranaldi F, Paoli P, Mugnaioni C, Michelucci E, Cirri P (2015) Cancer associated fibroblasts transfer lipids and proteins to cancer cells through cargo vesicles supporting tumor growth. Biochim Biophys Acta 1853(12):3211–3223. https://doi.org/10.1016/j.bbamcr.2015.09.013
Feng Y-H, Tsao C-J (2016) Emerging role of microRNA-21 in cancer. Biomed Rep 5(4):395–402. https://doi.org/10.3892/br.2016.747
Chen S, Chen X, Shan T, Ma J, Lin W, Li W, Kang Y (2018) MiR-21-mediated Metabolic Alteration of Cancer-associated Fibroblasts and Its Effect on Pancreatic Cancer Cell Behavior. Int J Biol Sci 14(1):100–110. https://doi.org/10.7150/ijbs.22555
Zhang H, Wang D, Li M, Plecita-Hlavata L, D'Alessandro A, Tauber J, Riddle S, Kumar S, Flockton A, McKeon BA, Frid MG, Reisz JA, Caruso P, El Kasmi KC, Jezek P, Morrell NW, Hu CJ, Stenmark KR (2017) Metabolic and Proliferative State of Vascular Adventitial Fibroblasts in Pulmonary Hypertension Is Regulated Through a MicroRNA-124/PTBP1 (Polypyrimidine Tract Binding Protein 1)/Pyruvate Kinase Muscle Axis. Circulation 136(25):2468–2485. https://doi.org/10.1161/circulationaha.117.028069
Wu H, Zhu S, Mo Y-Y (2009) Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res 19(4):439–448. https://doi.org/10.1038/cr.2009.18
Giannoni E, Taddei ML, Morandi A, Comito G, Calvani M, Bianchini F, Richichi B, Raugei G, Wong N, Tang D, Chiarugi P (2015) Targeting stromal-induced pyruvate kinase M2 nuclear translocation impairs oxphos and prostate cancer metastatic spread. Oncotarget 6(27):24061–24074. https://doi.org/10.18632/oncotarget.4448
Aird WC (2012) Endothelial Cell Heterogeneity. Cold Spring Harb Perspect Med 2(1):a006429. https://doi.org/10.1101/cshperspect.a006429
Lopes-Bastos BM, Jiang WG, Cai J (2016) Tumour-Endothelial Cell Communications: Important and Indispensable Mediators of Tumour Angiogenesis. Anticancer Res 36(3):1119–1126
Dudley AC (2012) Tumor Endothelial Cells. Cold Spring Harb Perspect Med 2(3):a006536. https://doi.org/10.1101/cshperspect.a006536
Azoitei N, Becher A, Steinestel K, Rouhi A, Diepold K, Genze F, Simmet T, Seufferlein T (2016) PKM2 promotes tumor angiogenesis by regulating HIF-1alpha through NF-kappaB activation. Mol Cancer 15:3. https://doi.org/10.1186/s12943-015-0490-2
Lin Y, Liu F, Fan Y, Qian X, Lang R, Gu F, Gu J, Fu L (2015) Both high expression of pyruvate kinase M2 and vascular endothelial growth factor-C predicts poorer prognosis in human breast cancer. Int J Clin Exp Pathol 8(7):8028–8037
Sun H, Zhu A, Zhang L, Zhang J, Zhong Z, Wang F (2015) Knockdown of PKM2 Suppresses Tumor Growth and Invasion in Lung Adenocarcinoma. Int J Mol Sci 16(10):24574–24587. https://doi.org/10.3390/ijms161024574
Ma T, Patel H, Babapoor-Farrokhran S, Franklin R, Semenza GL, Sodhi A, Montaner S (2015) KSHV induces aerobic glycolysis and angiogenesis through HIF-1-dependent upregulation of pyruvate kinase 2 in Kaposi’s sarcoma. Angiogenesis 18:477–488
Lee E, Pandey NB, Popel AS (2014) Lymphatic endothelial cells support tumor growth in breast cancer. Sci Rep 4:5853. https://doi.org/10.1038/srep05853 https://www.nature.com/articles/srep05853#supplementary-information
He Z, Yi J, Liu X, Chen J, Han S, Jin L, Chen L, Song H (2016) MiR-143-3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial–mesenchymal transition by targeting QKI-5 in esophageal squamous cell carcinoma. Mol Cancer 15(1):51. https://doi.org/10.1186/s12943-016-0533-3
Xu RH, Liu B, Wu JD, Yan YY, Wang JN (2016) miR-143 is involved in endothelial cell dysfunction through suppression of glycolysis and correlated with atherosclerotic plaques formation. Eur Rev Med Pharmacol Sci 20(19):4063–4071
Zhang L, Chen X, Liu B, Han J (2018) MicroRNA-124-3p directly targets PDCD6 to inhibit metastasis in breast cancer. Oncol Lett 15(1):984–990. https://doi.org/10.3892/ol.2017.7358
Caruso P, Dunmore BJ, Schlosser K, Schoors S, Dos Santos C, Perez-Iratxeta C, Lavoie JR, Zhang H, Long L, Flockton AR, Frid MG, Upton PD, D'Alessandro A, Hadinnapola C, Kiskin FN, Taha M, Hurst LA, Ormiston ML, Hata A, Stenmark KR, Carmeliet P, Stewart DJ, Morrell NW (2017) Identification of MicroRNA-124 as a Major Regulator of Enhanced Endothelial Cell Glycolysis in Pulmonary Arterial Hypertension via PTBP1 (Polypyrimidine Tract Binding Protein) and Pyruvate Kinase M2. Circulation 136(25):2451–2467. https://doi.org/10.1161/circulationaha.117.028034
Esteve Rafols M (2014) Adipose tissue: cell heterogeneity and functional diversity) Endocrinologia y nutricion : organo de la Sociedad Espanola de. Endocrinol Nutr 61(2):100–112. https://doi.org/10.1016/j.endonu.2013.03.011
Zahid H, Simpson ER, Brown KA (2016) Inflammation, dysregulated metabolism and aromatase in obesity and breast cancer. Curr Opin Pharmacol 31:90–96. https://doi.org/10.1016/j.coph.2016.11.003
Jiang Y, Guo L, Xie LQ, Zhang YY, Liu XH, Zhang Y, Zhu H, Yang PY, Lu HJ, Tang QQ (2014) Proteome profiling of mitotic clonal expansion during 3T3-L1 adipocyte differentiation using iTRAQ-2DLC-MS/MS. J Proteome Res 13(3):1307–1314. https://doi.org/10.1021/pr401292p
Wei L, Li K, Pang X, Guo B, Su M, Huang Y, Wang N, Ji F, Zhong C, Yang J, Zhang Z, Jiang Y, Liu Y, Chen T (2016) Leptin promotes epithelial-mesenchymal transition of breast cancer via the upregulation of pyruvate kinase M2. J Exp Clin Cancer Res : CR 35(1):166. https://doi.org/10.1186/s13046-016-0446-4
Liao SC, Li JX, Yu L, Sun SR (2017) Protein tyrosine phosphatase 1B expression contributes to the development of breast cancer. J Zhejiang Univ Sci B 18(4):334–342. https://doi.org/10.1631/jzus.B1600184
Bettaieb A, Bakke J, Nagata N, Matsuo K, Xi Y, Liu S, AbouBechara D, Melhem R, Stanhope K, Cummings B, Graham J, Bremer A, Zhang S, Lyssiotis CA, Zhang ZY, Cantley LC, Havel PJ, Haj FG (2013) Protein tyrosine phosphatase 1B regulates pyruvate kinase M2 tyrosine phosphorylation. J Biol Chem 288(24):17360–17371. https://doi.org/10.1074/jbc.M112.441469
Xie J, Dai C, Hu X (2016) Evidence That Does Not Support Pyruvate Kinase M2 (PKM2)-catalyzed Reaction as a Rate-limiting Step in Cancer Cell Glycolysis. J Biol Chem 291(17):8987–8999. https://doi.org/10.1074/jbc.M115.704825
Hosios AM, Fiske BP, Gui DY, Vander Heiden MG (2015) Lack of evidence for PKM2 protein kinase activity. Mol Cell 59(5):850–857
Cheng H, Qin Y, Fan H, Su P, Zhang X, Zhang H, Zhou G (2013) Overexpression of CARM1 in breast cancer is correlated with poorly characterized clinicopathologic parameters and molecular subtypes. Diagn Pathol 8:129–129. https://doi.org/10.1186/1746-1596-8-129
Kung C, Hixon J, Choe S, Marks K, Gross S, Murphy E, DeLaBarre B, Cianchetta G, Sethumadhavan S, Wang X (2012) Small molecule activation of PKM2 in cancer cells induces serine auxotrophy. Chem Biol 19(9):1187–1198
Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW, Burga LN, Xie J, Jurczak MJ, DePinho RA, Clish CB, Jacks T, Kibbey RG, Wulf GM, Di Vizio D, Mills GB, Cantley LC, Vander Heiden MG (2013) PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell 155(2):397–409. https://doi.org/10.1016/j.cell.2013.09.025
Cortes-Cros M, Hemmerlin C, Ferretti S, Zhang J, Gounarides JS, Yin H, Muller A, Haberkorn A, Chene P, Sellers WR, Hofmann F (2013) M2 isoform of pyruvate kinase is dispensable for tumor maintenance and growth. Proc Natl Acad Sci U S A 110(2):489–494. https://doi.org/10.1073/pnas.1212780110
Dayton TL, Gocheva V, Miller KM, Israelsen WJ, Bhutkar A, Clish CB, Davidson SM, Luengo A, Bronson RT, Jacks T (2016) Germline loss of PKM2 promotes metabolic distress and hepatocellular carcinoma. Genes Dev
Dayton TL, Gocheva V, Miller KM, Bhutkar A, Lewis CA, Bronson RT, Vander Heiden MG, Jacks T (2018) Isoform-specific deletion of PKM2 constrains tumor initiation in a mouse model of soft tissue sarcoma. Cancer & metabolism 6, 6(1)
Stetak A, Veress R, Ovadi J, Csermely P, Keri G, Ullrich A (2007) Nuclear translocation of the tumor marker pyruvate kinase M2 induces programmed cell death. Cancer research 67(4):1602–1608. https://doi.org/10.1158/0008-5472.can-06-2870
Hillis AL, Lau AN, Devoe CX, Dayton TL, Danai LV, Di Vizio D, Vander Heiden MG (2018) PKM2 is not required for pancreatic ductal adenocarcinoma. Cancer Metab 6(1):17
Lau AN, Israelsen WJ, Roper J, Sinnamon MJ, Georgeon L, Dayton TL, Hillis AL, Yilmaz OH, Di Vizio D, Hung KE (2017) PKM2 is not required for colon cancer initiated by APC loss. Cancer Metab 5(1):10
Curless BP, Uko NE, Matesic DF (2018) Modulator of the PI3K/Akt oncogenic pathway affects mTOR complex 2 in human adenocarcinoma cells. Investig New Drugs:1–10
Li Y, Bao M, Yang C, Chen J, Zhou S, Sun R, Wu C, Li X, Bao J (2018) Computer-aided identification of a novel pyruvate kinase M2 activator compound. Cell Prolif 51(6):e12509
Li R, Ning X, Zhou S, Lin Z, Wu X, Chen H, Bai X, Wang X, Ge Z, Li R (2018) Discovery and structure-activity relationship of novel 4-hydroxy-thiazolidine-2-thione derivatives as tumor cell specific pyruvate kinase M2 activators. Eur J Med Chem 143:48–65
Ning X, Qi H, Li R, Li Y, Jin Y, McNutt MA, Liu J, Yin Y (2017) Discovery of novel naphthoquinone derivatives as inhibitors of the tumor cell specific M2 isoform of pyruvate kinase. Eur J Med Chem 138:343–352
Zhang Y, Liu B, Wu X, Li R, Ning X, Liu Y, Liu Z, Ge Z, Li R, Yin Y (2015) New pyridin-3-ylmethyl carbamodithioic esters activate pyruvate kinase M2 and potential anticancer lead compounds. Bioorg Med Chem 23(15):4815–4823
Chen J, Xie J, Jiang Z, Wang B, Wang Y, Hu X (2011) Shikonin and its analogs inhibit cancer cell glycolysis by targeting tumor pyruvate kinase-M2. Oncogene 30(42):4297–4306. https://doi.org/10.1038/onc.2011.137
Palsson-McDermott EM, Dyck L, Zasłona Z, Menon D, McGettrick AF, Mills KH, O’Neill LA (2017) Pyruvate kinase M2 is required for the expression of the immune checkpoint PD-L1 in immune cells and tumors. Front Immunol 8:1300
Yacovan A, Ozeri R, Kehat T, Mirilashvili S, Sherman D, Aizikovich A, Shitrit A, Ben-Zeev E, Schutz N, Bohana-Kashtan O (2012) 1-(sulfonyl)-5-(arylsulfonyl) indoline as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorg Med Chem Lett 22(20):6460–6468
Guo C, Linton A, Jalaie M, Kephart S, Ornelas M, Pairish M, Greasley S, Richardson P, Maegley K, Hickey M (2013) Discovery of 2-((1H-benzo [d] imidazol-1-yl) methyl)-4H-pyrido [1, 2-a] pyrimidin-4-ones as novel PKM2 activators. Bioorg Med Chem Lett 23(11):3358–3363
Xu Y, Liu X-H, Saunders M, Pearce S, Foulks JM, Parnell KM, Clifford A, Nix RN, Bullough J, Hendrickson TF (2014) Discovery of 3-(trifluoromethyl)-1H-pyrazole-5-carboxamide activators of the M2 isoform of pyruvate kinase (PKM2). Bioorg Med Chem Lett 24(2):515–519
Walsh MJ, Brimacombe KR, Anastasiou D, Yu Y, Israelsen WJ, Hong B-S, Tempel W, Dimov S, Veith H, Yang H (2013) ML265: A potent PKM2 activator induces tetramerization and reduces tumor formation and size in a mouse xenograft model. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. National Center for Biotechnology Information (US)
Brimacombe KR, Anastasiou D, Hong B-S, Tempel W, Dimov S, Veith H, Auld DS, Vander Heiden MG, Thomas CJ, Park H-W (2013) ML285 affects reactive oxygen species’ inhibition of pyruvate kinase M2.
Kim DJ, Park YS, Kim ND, Min SH, You Y-M, Jung Y, Koo H, Noh H, Kim J-A, Park KC (2015) A novel pyruvate kinase M2 activator compound that suppresses lung cancer cell viability under hypoxia. Mol Cells 38(4):373
Porporato PE, Dhup S, Dadhich RK, Copetti T, Sonveaux P (2011) Anticancer targets in the glycolytic metabolism of tumors: a comprehensive review. Front Pharmacol 2:49–49. https://doi.org/10.3389/fphar.2011.00049
Martins P, Jesus J, Santos S, Raposo L, Roma-Rodrigues C, Baptista P, Fernandes A (2015) Heterocyclic anticancer compounds: recent advances and the paradigm shift towards the use of nanomedicine’s tool box. Molecules 20(9):16852–16891
Spilioti E, Jaakkola M, Tolonen T, Lipponen M, Virtanen V, Chinou I, Kassi E, Karabournioti S, Moutsatsou P (2014) Phenolic acid composition, antiatherogenic and anticancer potential of honeys derived from various regions in Greece. PloS One 9(4):e94860
Acknowledgments
We acknowledge the support from National Institute of Pharmaceutical Education and Research (NIPER), Ahmedabad.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rihan, M., Nalla, L.V., Dharavath, A. et al. Pyruvate Kinase M2: a Metabolic Bug in Re-Wiring the Tumor Microenvironment. Cancer Microenvironment 12, 149–167 (2019). https://doi.org/10.1007/s12307-019-00226-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12307-019-00226-0