Abstract
The initial components of ribosomes first appeared more than 3.8 billion years ago during a time when many types of RNAs were evolving. While modern ribosomes are complex molecular machines consisting of rRNAs and proteins, they were assembled during early evolution by the association and joining of small functional RNA units. Introns may have provided the means to ligate many of these pieces together. All four classes of introns (group I, group II, spliceosomal, and archaeal) are present in many rRNA gene loci over a broad phylogenetic range. A survey of rRNA intron sequences across the three major life domains suggests that some of the classes of introns may have diverged from one another within rRNA gene loci. Analyses of rRNA sequences revealed self-splicing group I and group II introns are present in ancestral regions of the SSU (small subunit) and LSU (large subunit), whereas spliceosomal and archaeal introns appeared in sections of the rRNA that evolved later. Most classes of introns increased in number for approximately 1 billion years. However, their frequencies are low in the most recently evolved regions added to the SSU and LSU rRNAs. Furthermore, many of the introns appear to have been in the same locations for billions of years, suggesting an ancient origin for these sequences. In this Perspectives paper, I reviewed and analyzed rRNA intron sequences, locations, structural characteristics, and splicing mechanisms; and suggest that rRNA gene loci may have served as evolutionary nurseries for intron formation and diversification.
Similar content being viewed by others
Introduction
From approximately 4.2–3.8 billion years ago, many innovative types of RNA evolved that played major roles in biology, during an era known as the “RNA world” (Darnell et al. 1990; Alberts et al. 1998; Gilbert et al. 1997; Wekselman et al. 2009; Belousoff et al. 2010; Rogers 2017; Bada 2013; Huang et al. 2013; Iwasa and Marshall 2016). This led to the assembly of the essential components of the central metabolic, evolutionary, and translational functions of ancient cells that still exist in modern cells (Darnell and Doolittle 1986; Gilbert et al. 1997; Bujnicki and Rychlewski 2001; Roy et al. 2002; Bokov and Steinberg 2009; Rogers 2017; Iwasa and Marshall 2016). They included catalytic RNAs (or ribozymes), structural RNAs, many classes of small RNAs, and functional nucleotide-containing compounds. Many of these compounds are still present in contemporary cells, such as ATP, GTP, NADH, hammerhead ribozyme, hairpin ribozyme, RNase P, small nucleolar RNAs (snoRNAs), rRNAs (ribosomal RNAs), tRNAs (transfer RNAs), mRNAs (messenger RNAs), introns, and others. Although pieces of what would become portions of rRNAs and ribosomes first appeared from 3.8 to 4.2 billion years ago, the first rRNAs that contained the PTC (peptidyltransferase center), and therefore, could synthesize small polypeptides, appeared approximately 3.6 to 3.8 billion years ago (Petrov et al. 2014, 2015; Fig. 1). However, these early ribosomes also relied on the delivery of amino acids via tRNAs, which were charged by aminoacyl tRNA synthetases (protein enzymes), and short mRNAs. It is clear that, from very early in their evolution, ribosomes were (and still are) composed of complexes of structural, enzymatic, kinetic, and binding RNAs, as well as structural proteins (Bokov and Steinberg 2009; Fox 2010; Huang et al. 2013; Petrov et al. 2014, 2015; Caetano-Annolés et al. 2013; Caetano-Annolés and Caetano-Annolés 2015; Rogers 2017).
Evolution of components of translation (including rRNA, tRNA, and mRNA). Six phases (top; Petrov et al. 2014, 2015), beginning more than 3.8 bya, are indicated for the evolution of AARSs (A, B), tRNA (C), mRNA (D), SSU rRNA (E), LSU rRNA (F). Colors indicate stage of expansions of the RNAs; from most ancient to most recent: blue, light blue, green, yellow, olive, and red
Many studies of ribosomes suggest that they were built from separate components, which were added sequentially over time (Fig. 1), and that the genetic code emerged in one of the later phases of rRNA evolution (Darnell and Doolittle 1986; Gilbert et al. 1997; Roy et al. 2002; Bokov and Steinberg 2009; Fujishima and Kanai 2014; Petrov et al. 2014, 2015; Root-Bernstein and Root-Bernstein 2015; Rogers 2017). Recent studies indicate that sections of both the rRNA large subunit (LSU) and small subunit (SSU) predated the PTC of the LSU, and that the two subunits evolved separately prior to their association to form a protoribosome (Bokov and Steinberg 2009; Petrov et al. 2014, 2015). For example, the PTC, tunnel, subunit association sites, P site (which holds the tRNA with the growing polypeptide), A site (which holds the tRNA with the incoming amino acid), and E site (where the exiting uncharged tRNA dissociates from the ribosome) were incorporated into rRNA at different times during early ribosome evolution, and complex decoding emerged only after all of these elements had been incorporated into the rRNA (Rodin and Rodin 2008; Bokov and Steinberg 2009; Wekselman et al. 2009; Belousoff et al. 2010; Fox 2010; Harish and Caetano-Anollés 2012; Caetano-Anollés et al. 2013; Huang et al. 2013; Petrov et al. 2014, 2015; Caetano-Anollés and Caetano-Anollés 2015). While the rRNA PTC appeared in one of the early phases of rRNA evolution, the coding and refinement of the genetic code by ribosomes evolved hundreds of millions of years later (Petrov et al. 2014, 2015). The appearance of the PTC adjacent to the P site and tunnel that became incorporated into the protoribosome approximately 3.6–3.8 billion years ago ushered in the initial stages of modern protein synthesis. The A site may have originated as a duplication and ligation of the P site sequence (Bokov and Steinberg 2009), as both have structural similarities and each is approximately 110 nt in length. Prior to that time the less efficient process of non-ribosomal protein synthesis (NRPS) was active more than 3.8 billion years ago. While NRPS is still present in contemporary cells, it is a minor process (which produces only peptides) compared to the translation of proteins by ribosomes. Translation by ribosomes became more efficient and more accurate than NRPS, and became the predominant method for protein synthesis.
A crucial part in the evolution of the protoribosome was that of ligation of the component pieces of RNA. Introns are plausible candidates for these RNA ligation, splicing, and alternative splicing reactions; and ligation continues to be one of their major functions. Many still exist within rDNAs (rRNA genes) of bacterial and archaeal species, as well as in nuclear and organellar genomes of a wide diversity of eukaryotes (Figs. 2, 3, 4), many being possible remnants of ancient ligation processes (Bhattacharya et al. 1996; Itoh et al. 1998; Jackson et al. 2002; Hackett et al. 2004; Haugen et al. 2005; Edgell et al. 2011; Moriera et al. 2012). The number and phylogenetic breadth of these introns suggest an ancient origin in the rRNA gene loci, and a potential coupling of evolutionary pathways of rRNAs and introns. In this Perspectives paper, it is proposed that introns have played a vital and central role in the evolution and function of rRNA and ribosomes, by joining together functional RNAs sequentially in the evolutionary construction of rRNAs. Rather than being primarily disruptive elements (as commonly perceived), introns may have played significant roles from the initial stages of rRNA and ribosome evolution by ligating RNAs together. Introns have been found in the most ancient parts of rRNAs, suggesting primeval origins (Jackson et al. 2002; Petrov et al. 2014, 2015). This, together with reports that rRNAs were formed from the sequential addition of portions of rRNA during their early evolution, suggests that introns may have played a role in piecing together the functional sections of RNA that later became the rRNAs and, ultimately, ribosomes (Bokov and Steinberg 2009; Wekselman et al. 2009; Belousoff et al. 2010; Huang et al. 2013; Petrov et al. 2014, 2015).
Taxonomic distribution and evolutionary pathways of group I (GrI), group II (GrII), spliceosomal (Spl), and archaeal (Arch) introns among Domains, Kingdoms, Phyla, and organelles. The tree is based on a consensus of several phylogenetic trees (Baldaus 2003; Roger and Simpson 2009; Hug et al. 2016). Solid lines indicate vertical inheritance of introns, while dashed lines with arrow heads indicate horizontal gene transfers via known endosymbiotic events (long dashes indicate primary endosymbiotic transfers, while short dashes indicate secondary and tertiary endosymbiotic transfers). Endosymbiotic events are indicated by black squares enclosing white letters (see Results for description of endosymbiotic events) Processing of rDNA in Archaea and Bacteria resembles archaeal intron splicing, and thus “Arch” is in parentheses for Bacteria. For Eukarya, white rounded rectangles indicate introns within mitochondrial genomes and green ovals indicate introns within photosynthetic organelle genomes. Green ovals with dashed borders indicate organelles that are present only in a few members of the taxon, although organellar sequences are present in the nuclear genomes. All other labels indicate introns in the genomes of Bacteria, Archaea, or nuclear genomes in Eukarya
Known locations of introns in SSU (top) and LSU (bottom) rRNA. Group I, group II, spliceosomal, and archaeal introns are represented by red, green, blue, and black arrows and nucleotide position numbers, respectively. Intron positions in the SSU rRNA (top) are relative to those in the E. coli SSU rRNA. Domains I (also called the 5′ region), II (also called the center, or C, region), III (also called the 3′ Major, or 3′M, region) and IV (also called the 3′ minor, or 3′m, region) are indicated. Positions in the LSU rRNA (bottom) are relative to those in the Saccharomyces cerevisiae LSU rRNA. Domains I, II, III, IV, V, and VI are indicated. The PTC is shown within domain V (dashed red circle). SSU and LSU rRNA colors are as in Fig. 1
In addition to the accretion of RNA pieces over billions of years, rRNA genes appear to have become nurseries for introns, functioning as evolutionary crucibles, which has resulted in the evolution and diversification of the four known classes of introns. This may be due to the fact that rRNA genes are present in multiple copies in most organisms (up to more than a dozen copies per genome in Bacteria and Archaea, and up to tens of thousands of copies per nuclear genome in Eukarya), they are some of the most transcriptionally active loci in cells, and they undergo frequent recombination and gene conversion events (Rogers et al. 1986; Rogers and Bendich 1987a, b). A large number of introns of all four classes have been described in rRNA genes from a broad phylogenetic range of organisms and organelles (Jackson et al. 2002), consistent with an ancient origin for these introns.
Together, the characterizations of rRNAs across the three major domains of life indicates that the introns and rRNAs have had long and coupled evolutionary histories, which have had important influences on the evolution of genomes, organisms, and organelles. The analyses presented here suggest that: (1) The phylogenetic breadth and diversity in the four intron classes point to a very early and sustained evolutionary history within rRNAs; (2) Introns of different classes often physically overlap each other in location and characteristics, suggesting connected evolutionary histories, including conversions from one class to another; (3) Small degenerate introns rely on portions of the rRNAs to complement missing intron core regions, demonstrating similarities in functions among the introns and rRNAs, the functional flexiblities of the RNAs, and their integrated evolutionary pathways; and (4) The number and diversity of introns within rRNA loci suggest that rRNAs may serve as intron nurseries that are sources for introns in other genomic regions. These results suggest the coupling of ribosomal RNA and intron evolution.
Methods
Evolution of translation components
Figure 1 is a summary of the evolution of several of the central components of translation based on the review of multiple studies on RNA evolution. This figure provides an overview of the major steps in the translation machinery. Information regarding the evolution of aminoacyl-tRNA synthetases was from Roberts et al. (2008, 2011), and Havrylenko and Mirande (2015). The steps in tRNA evolution are from Fujishima and Kanai (2014), and Kanai (2015). The evolution of mRNA, including the early use of double stranded mRNAs, is from Rodin and Rodin (2008), Rodin et al. (2011), and Root-Bernstein and Root-Bernstein (2015). Evolution of the SSU and LSU rRNAs and dates are based on Petrov et al. (2014, 2015).
Phylogenetic and sequence distribution of introns
A comprehensive collection of introns within rDNA (Rogers et al. 1993; Gargas et al. 1995; Shivji et al. 1995; Bhattacharya et al. 1996, 2001; Itoh et al. 1998; Jackson et al. 2002; Haugen et al. 2005; Moriera et al. 2012) from a broad diversity of organisms was mapped onto a SSU rRNA tree, constructed as a consensus tree among a collection of phylogenetic trees (Baldauf 2003; Roger and Simpson 2009; Hug et al. 2016; see Fig. 2). This tree indicates the taxonomic distribution and mode of transfer (vertical versus horizontal acquisition) of the four different classes of introns analyzed.
The introns were also examined according to their location in rRNA sequences and when they appeared during the six major phases of rRNA evolution (Petrov et al. 2014, 2015; Fig. 3). Introns of all four classes were mapped onto the SSU and LSU rRNAs, relative to their positions in the E. coli SSU rRNAand Saccharomyces cerevisiae LSU rRNA (Davila-Aponte et al. 1991; DeWachter et al. 1992; Gast et al. 1994; Murphy and Cech 1994; Gargas et al. 1995; Bhattacharya et al. 1996, 2000, 2003; Grube et al. 1996; Shinohara et al. 1996; Chen et al. 1998; Itoh et al. 1998; Perotto et al. 2000; Nikoh and Fukatsu 2001; Cannone et al. 2002; Edgell et al. 2000; Jackson et al. 2002; Lickey et al. 2003; Haugen et al. 2004; Gutierrez et al. 2007; Harris 2007; Harris and Rogers 2008, 2011; Chen 2010; Salman et al. 2012; Petrov et al. 2014, 2015). The frequencies of each class of intron in each of the regions were then compared relative to each of the six phases of ribosomal evolution (Fig. 4).
Structural characterization of introns
Analyses of the secondary structure of group I and group II proteins was performed using Mfold (Zucker 2003) to identify structural homologies on introns’ splice/insertion sites and thus demonstrate the conversion and functional flexibility of the introns and the rRNA.
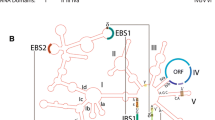
Source cultures, extraction of DNA, PCR analyses, sequencing, cloning, descriptions, categorization, phylogenetic position, mutant intron synthesis, in vitro transcription, and splicing assays for the CgSSU (from Ceonococcum geophilum SSU rDNA) and PaSSU (from Phialophora americana SSU rDNA) introns, both at rRNA nucleotide (nt) position 1506, are described elsewhere (Harris 2007; Harris and Rogers 2008, 2011, 2014; Rogers et al. 1993; Shinohara et al. 1996; Yan et al. 1995). The sequences of the CgSSU and PaSSU introns, both inserted into the 1506 position of the SSU rRNA genes (Rogers et al. 1993), were subjected to group I intron secondary structure analysis using Mfold (Zuker 2003). Because of the length of the CgSSU intron (459 nt), structures of this intron were made in overlapping sections less than 350 nt in length. The sections were overlapped and joined for consistency (Fig. 5, top). The PaSSU intron structure was determined by a single analysis, because of its small size (67 nt; Fig. 6). To produce the final structure, manual adjustments for both structures were made based on maximization of base pairing and ΔG, while considering short and long range interactions based on previous group I intron models (Adams et al. 2004; Cech 1990; Michel and Westof 1990; Rogers et al. 1993; Shinohara et al. 1996). Both introns had a branch sequence near the 3′ end of the intron (Fig. 7) that is characteristic of group II introns. Because of this, the CgSSU intron sequence was subjected to group II intron structure analysis (Fig. 5, bottom), although the splice/insertion site was shifted by -2 nt (position 1504 in the SSU rRNA), where a characteristic group II splicing sequence was located. The focus was on the potential group II intron structures, based on published group II models (Michel et al. 1989; Seetharaman et al. 2006). This analysis was performed to determine whether a group I and a group II intron were coincident at this site.
Structure of a group I intron conformation (top) and group II intron conformation (bottom) for the same intron sequence within the 3′ region (positions 1506 and 1504, respectively) of the SSU rRNA from the fungus Cenococcum geophilum. Nucleotides within the intron are in black upper case font, while the 5′ and 3′ exons are in red lower case font. For the group I intron (top), the intron/exon (I/E) borders are indicated with filled arrows. Intron/exon borders for a spliceosomal or group II mechanism are indicated with open dashed arrows. An alternative structure (bottom) of the Cenococcum geophilum SSU rRNA intron sequence at the same position (position 1504) as the group I intron is folded into a group II intron structure. Six domains (D1 through D6) are indicated
Results of mutational and in vitro splicing assays for the PaSSU degenerate group I intron (67 nt) from Phialophora americana. Several cloned constructs were synthesized that included the small intron and parts of the 5′ and/or 3′ exons. Upper case fonts indicate nucleotides within the intron, while lower case fonts indicate nucleotides within the exons. Mutations that resulted in reduced splicing are highlighted in red, while those that had little or no effect on splicing are indicated in green
Results of in vitro splicing of the PaSSU degenerate group I intron (67 nucleotides) from Phialophora americana, with surrounding exons in the SSU rRNA. The upper portion represents the results from group I intron experimental results, while the lower portion indicates the portions of the sequences that are in common with group II intron splicing. The solid lines in the upper portion indicate the major splice sites, while the dashed lines indicate the minor splice sites. The lower dotted lines indicate the splicing borders for a spliceosomal (or group II) splicing mechanism. The conserved branch site for a spliceosomal (or group II) intron is indicated by letters underlined in bold within the intron. The internal A that initiates splicing is indicated by an arrow
Mutant intron splicing assays
In addition to the structural characterization of selected introns, a series of in vitro splicing assays of mutant introns from P. americana were performed to evaluate/assess the functional role of specific regions of the rRNA on the splicing process (methods and results are described in detail in Harris 2007; and; Harris and Rogers 2008, 2010, 2014). The PaSSU degenerate intron (67 nt) from P. americana, including approximately 200 bp of the SSU rDNA upstream and approximately 200 bp of the downstream SSU, ITS1, and 5.8S rRNA, was cloned into an expression vector to produce RNA for splicing assays. In vitro splicing assays were performed to determine whether portions of the rRNA were required for splicing of the PaSSU degenerate intron (Fig. 6). Mutant clones were constructed separately by deleting the 5′ upstream sections and 3′ downstream sections, as well as changing, deleting, or inserting selected nucleotides within the intron to examine the effects on splicing.
Results
Phylogenetic distribution of introns
The cladogram of rRNA SSU sequences (Fig. 2) reveals that many members of the four classes of introns have been inherited vertically during evolutionary processes, while others have been transferred horizontally from one species to another via endosymbiotic events, horizontal gene transfers, or have migrated from one cell compartment to another (Bhattacharya et al. 2001; Hackett et al. 2004; Haugen et al. 2005; Moriera et al. 2012; Rogers 2017). Endosymbiotic events indicative of horizontal transfer included: formation of mitochondria from an alphaproteobaceterium in Eukarya (Fig. 2, box a); formation of plastids from a cyanobacterium in Archaeplastida (Fig. 2, box b); formation of a cyanelle from a cyanobacterium in Rhizaria (Fig. 2, box c); formation of a photosynthetic organelle from a red alga (Rhodophyta) in the Stramenopile, Alveolata, Rhizaria, and Hacrobia clade (Fig. 2, box d); formation of an intracellular organelle from a green alga (Chlorophyta) in Rhizaria (Fig. 2, box e); formation of an intracellular organelle from a green alga in Excavata (Fig. 2, box f); formation of a photosynthetic organelle from a Stramenopile in Alveolata (Fig. 2, box g); formation of an intracellular organelle from a member of the Alveolata in another member of Alveolata (Fig. 2, box h); and formation of an intracellular organelle from a member of the Hacrobia in Alveolata (Fig. 2, box i).
Processing of rDNA in Archaea and Bacteria resembles archaeal intron splicing. This is indicated in Fig. 2 as “Arch” in parentheses for Bacteria (which otherwise have no known archaeal introns). Archaeal intron derivatives may be present in the internal transcribed spacers of nuclear and organellar genomes, but these are not shown.
The most parsimonious position for spliceosomal introns is their derivation from group II introns during the origin of Eukarya (approximately 2.4 billion years ago; shown by an arrow early in eukaryote evolution, near the Excavata branch). However, their presence in more ancient portions of the rRNAs, suggests that they may have had an earlier origin.
The local positioning of introns in rRNAs was assessed by mapping all four classes of introns onto the SSU and LSU rRNAs (Fig. 3). Approximately equal numbers of introns are inserted into single stranded regions (41) as in double stranded regions (39) in the SSU rRNAs, whereas more of the introns are located in single stranded regions (44) than in double stranded regions (34) in the LSU rRNAs. Approximately 33% of the introns in the SSU rRNAs and 25% of the introns in the LSU rRNAs were within 10 nt of the borders between more ancient and more recently evolved sections of the SSU rRNAs. Furthermore, only group I and group II introns were located in the PTC of the LSU rRNAs (Fig. 3).
Appearance of the introns during evolution
The examination of introns and their frequency distributions in relation to the six stages of ribosomal evolution provided insights into the early evolution of the four classes of introns. Group I introns were present in regions of the first phase of SSU rRNA evolution (Figs. 1, 3, 4), and increased in frequency into phase 2, decreased slightly in phase 3, and increased greatly in phases 4 and 5, followed by a sharp decrease during phase 6. Group II introns were first present in rRNA phase 2, increased slightly, then decreased, coinciding with a rapid increase in spliceosomal introns during phases 3, 4, and 6, with a notable decrease during phase 5. Archaeal introns appeared first in rRNA phase 2, and increased and decreased in frequency during the subsequent phases. In the LSU rRNA, group I, group II, and spliceosomal introns were located in the most ancient regions, although in low frequencies. Increases in group I and group II introns occured in phase 2, but spliceosomal introns are not found in the same regions. It is possible that the splieosomal introns found in phase 1 regions are more recent conversions from group II introns. As with the SSU rRNA, there were decreases in most intron classes (except spliceosomal introns, which increase), and subsequent increases of group I, group II, and spliceosomal introns, during phases 4 and 5. No introns were present in the most recently evolved regions of the LSU rRNA (phase 6). Archaeal introns occurred more frequently in SSU rRNA genes than in LSU rRNA genes (Fig. 4).
Structural analyses
The structure of the group I intron in the SSU rRNA in the 1506 position is well established, and is clearly delineated in Fig. 5 (top). Pairing regions P1 through P10 are present. A characteristic G·U pair is present at the 5′ exon/intron splice site, and a guanosine binding site, which is a G-C pair with an adjacent unpaired A, is present in P7. The P4-P5-P6 domain acts as the scaffold of the intron. P2 is an optional stem-loop. The P3-P7-P8-P9 domain forms the catalytic arm of the intron that includes the guanosine binding site in P7. GNRA loops in P2, P5, and P9 are indicated in upper case red font. These are important in long range interactions. Joining regions J3/4, J6/7, and J7/8 (the most conserved of the three) also are shown. These are important in long range interactions. The CgSSU and PaSSU sequences also have similarities to group II and spliceosomal introns, in that group II exon–intron borders are located only 1–2 nucleotides in the 5′ direction from the group I border sites, and they have sequences that are similar to a typical group II branch site sequences a short distance from the 3′ intron–exon border (Figs. 6, 7). When the CgSSU sequence was subjected to structural analysis using Mfold (Zucker 2003), both group I and group II structures resulted (Fig. 5, top and bottom, respectively). The group II structure included the 5′ and 3′ splice sites, internal and external binding sites (IBS1, IBS2, EBS1, and EBS2), and stem-loop domains D1 through D6, including the D5 stem-loop that is universally conserved among group II introns (which contains the canonical central CA bulge and GAAA loop). The structural analysis showed that three additional GNRA loops and three GNRA bulges were also present (Fig. 5, lower portion). Also present (in blue font, and within boxes) are regions involved in long range interactions (ε and ε’, γ and γ’, and ξ and ξ’).
In vitro splicing experiments
To assess the potential role of different introns and rRNA sites and regions in the splicing process, the wild type PaSSU degenerate group I intron (67 nt in length) and several mutant recombinant constructs were subjected to in vitro group I splicing assays (Fig. 6; Harris 2007; Harris and Rogers 2008, 2011, 2014). Splicing occurred whether or not 200 nt of the 5′ exon (within the SSU rRNA) was present. When nucleotides in or near the 5′ splice site were changed (G-2, U-1, G1, or G15), splicing was negatively affected. Changing a single A residue in the A-loop of P1 (A7) also caused a cessation of splicing, while changing an unpaired U residue within P1 (U4) had no effect on splicing. Changes in pairing region P9 (which is suspected of substituting for P7 in this degenerate intron) also had mainly detrimental effects on splicing (e.g., changes to C43, G44, or U45), although changes in nucleotides that appeared to have no interactions with other nucleotides (e.g., G41 and G57) had no effect on splicing. Splicing using RNA from the clone with only 20 nt of each exon failed to splice in vitro in any of the numerous attempts. However, when clones included the 3′ exon (that consisted of ITS1 and the 5′ one-third of the 5.8S rRNA), splicing occurred, whether or not the 5′ exon was present. In contrast, when the 3′ exon (including ITS1 and the 5′ one-third of the 5.8S rRNA) was deleted, splicing was never observed (Harris 2007; Harris and Rogers 2008, 2011, 2014).
Discussion
This study shows several lines of evidence that indicate there are ancient links in the evolutionary histories of ribosomes and introns. Aside from the fact that both have ribozyme functions, they appear to have evolved intertwined for billions of years. First, the ribosomal RNA gene loci of a broad phylogenetic diversity of organisms and organelles contain introns in a variety of locations, and some have been described in the most ancient regions of rRNAs (Rogers et al. 1993; Gast et al. 1994; Johansen and Vogt 1994; Shivji et al. 1995; Bhattacharya et al. 1996b, 2000; Takai and Horikoshi 1999; Perotto et al. 2000; Muller et al. 2001; Cannone et al. 2002; Jackson et al. 2002; Roy et al. 2002; Haugen et al. 2004; Moreira et al. 2012; Rogers 2017; see Figs. 1, 2, 3, 4). In fact, many of the introns that are present in rRNAs today may be remnants of those that existed during the early evolution of rRNAs (although some are likely later additions). Second, all four classes of introns (group I, group II, spliceosomal, and archaeal) have been reported in rRNA genes (see Figs. 2, 3, 4). This differs from all other classes of genes, including nuclear mRNA genes, which contain only spliceosomal introns, and tRNA genes, which contain primarily archaeal introns. Again, this suggests an early evolutionary origin for these introns, and possibly a common origin for all four intron classes. Third, the sections of rRNA appear to have been joined together in sequential stages throughout rRNA evolution, rather than having expanded in a slow incremental fashion (Rodin and Rodin 2008; Bokov and Steinberg 2009; Fox 2010; Petrov et al. 2014, 2015; Rogers 2017; Figs. 3, 4). The main characteristic of introns is that they join pieces of RNA together, and thus are candidates for the process of joining sections of rRNA together. However, they also can insert into DNA genes, similar to transposons (for group I introns) or retrotransposons (for group II introns) and, therefore, may have introduced novel sections of rRNA genes by these mechanisms (Fig. 8). Furthermore, introns may also provide opportunities for unequal crossover events, which can lead to novel exon combinations (Rogers and Bendich 1987a, b, 1988; Rogers 2017). Fourth, processing of pre-rRNAs has many similarities to archaeal intron splicing, including the formation of initially circular products (Kjems and Garrett 1991; Belfort and Weiner 1997; Lykke-Anderson et al. 1997; Takai and Horikoshi 1999; Tang et al. 2002; Fujishima and Kanai 2014). This supports the thesis that archaeal introns have had a long history within the rRNA locus, and may have been responsible for joining some pieces of rRNA together, including joining the SSU and LSU rRNA genes together to generate the rRNA operon, as well as participating in pre-rRNA processing. Therefore, introns and rRNAs appear to have had long integrated evolutionary histories, and introns appear to have been intimately involved in the assembly and processing of ribosomal RNAs from their earliest appearance as small rudimentary RNAs more than 3.8 billion years ago.
Proposed model of the evolution of introns with rRNAs. Very early in rRNA evolution only RNAs were involved in the process (top). Introns were probably capable of ligating RNAs internally (left) or externally (right). Once the shift had been made to DNA genomes, the introns remained in the genes for longer periods of time, or they were lost, translocated, or duplicated. New introns inserted, and those already present often moved or inserted copies elsewhere in the same locus or in different loci. They mutated, increased, or decreased in size, or occasionally converted to a different type of intron. Once the loci were present in multiple copies, these processes could increase in frequency, thus becoming nurseries for introns
The results presented here also show an intricate relationship between the evolution of the LSU and SSU rRNA subunits and introns. The locations of introns in the LSU and SSU rRNAs are concentrated in the more ancient portions of those molecules, and few have been reported in the most recently evolved sections of rRNAs (Figs. 3, 4). For example, many introns are located within LSU rRNA domains II, IV, and V. These are among the earliest appearing sections of rRNA, which include the PTC, the P site, the A site, and the tunnel (Bokov and Steinberg 2009; Wekselman et al. 2009; Belousoff et al. 2010; Fox 2010; Huang et al. 2013; Petrov et al. 2014, 2015). Interestingly, only group I and group II introns occur in the PTC, which is suggestive of an ancient origin and rRNA association for these two classes of introns. Also, the PTC contains the highest concentrations of group II introns. Some group II introns move as RNAs rather than DNAs, which might have facilitated insertion early in rRNA evolution. No introns have been reported in LSU domains I, III, or VI, which contain higher proportions of sections that were later additions to the LSU rRNAs. Similarly, for the SSU rRNAs, the introns are concentrated in the more ancient central sections of the molecule, and in low frequency in the more recently lengthened stem-loop regions.
Previous studies have shown that the SSU and LSU ribosomal subunits initially evolved separately, and the SSU may have predated the LSU as an RNA binding molecule (Petrov et al. 2014, 2015; Caetano-Annolés and Caetano-Annolés 201; Fig. 1). The LSU appears to have begun with the PTC, the tunnel, and a rudimentary P site, which consisted of approximately 110 nucleotides (Bokov and Steinberg 2009; Petrov et al. 2014, 2015). The P site was duplicated to form the adjacent A site that may have been the result of the ligation of the two RNAs (Darnell and Doolittle 1986; Gilbert et al. 1997; Bokov and Steinberg 2009; Petrov et al. 2014, 2015). This likely led to increased efficiency of polypeptide synthesis, because the incoming tRNA at the A site would be held close to the tRNA in the P site and the PTC. The tunnel in the LSU rRNA, which is adjacent to the PTC, P site, and A site, also had an early origin (Bokov and Steinberg 2009; Wekselman et al. 2009; Belousoff et al. 2010; Fox 2010; Huang et al. 2013; Petrov et al. 2014, 2015). It helps to hold the growing polypeptide to the ribosome, so that the next amino acid can be added, and prevents circularization of the lengthening polypeptide. These are among the earliest sections of the LSU rRNA and contain a large number of introns, including all four types, although group I introns are the most numerous (Figs. 3, 4). Other portions of the rRNAs may have been added as preformed functional components, rather than by extension of existing rRNA sections. Again, this suggests that the sections of rRNA may have been ligated as functional components over time. This may have occurred via alternative splicing, promiscuous splicing, trans-splicing, transposition, insertion, unequal crossing over within the intron regions, or by other mechanisms.
Introns in ribosomal RNA
Hundreds of introns of all four types have been reported within rRNA gene loci among a large collection of eukaryotes (Jackson et al. 2002; Figs. 2, 3), and at least two types have been found in the rRNA gene loci of Bacteria (group I and group II), organelles (group I and group II), and Archaea (group I, group II, and archaeal). The frequency and diversity of these introns is notable (Figs. 3, 4), with many common positions within Bacteria and Archaea (e.g., SSU rRNA positions 299, 330, 393, 516, 788, 908, 940, 943, 1046, 1199, 1210, 1506, 1512, and 1516, common in a wide diverse group of organisms, Jackson et al. 2002; and positions 781, 1205, and 1213, within Thermoproteus species; Itoh et al. 1998). Many of these introns may have existed in the same locations for billions of years, as indicated by their phylogenetic relationships among taxonomic groups or within ancient organelles (e.g., mitochondria, plastids, and nuclei) that have existed for billions of years. For example, common intron positions 788 and 1210 are present within mitochondrial and nuclear SSU rRNA; positions 531 and 793 in mitochondrial and plastid SSU rRNA; positions 787 and 1249 in mitochondrial and nuclear LSU rRNA; positions 1065, 1951, 2500, and 2593 in mitochondrial and plastid LSU rRNA; positions 1923, 1931, and 2449 in mitochondrial, plastid, and nuclear LSU rRNA; and common intron sites in the nuclear rRNA in a diversity of fungi (Jackson et al. 2002; Rogers et al. 1993; Gargas et al. 1995; Shivji et al. 1995; Bhattacharya et al. 1996, 2001; Haugen et al. 2005; Moriera et al. 2012; Figs. 3, 4). Group I introns are also common in the plastid, mitochondrial, and nuclear rRNAs of green algae and plants, as well as in the mitochondrial and nuclear rRNAs of fungi and amoebae. Introns at positions 516 and 943 have been described from a number of green algae, amoebae, and fungi, and are phylogenetically closer to one another than they are to other introns within the same genetic locus, suggesting that they have existed in those locations since before those organisms diverged from a common ancestor more than 2.0 billion years ago (Gargas et al. 1995).
The importance of introns in evolution is indisputable, being the raw materials for transcript splicing, alternative splicing, fusion genes/proteins, certain noncoding RNAs, gene expression control, recombination sites, and others (Rogers 2017). They have been found in mRNA genes, tRNA genes, and rRNA genes, often in multiple locations in each gene. Furthermore, some introns contain genes for reverse transcriptases, endonucleases, resolvases, and noncoding RNAs, some of which confer mobility, while others exhibit wide ranging genetic, epigenetic, and phenotypic effects. While many genes contain no introns, some have more than a dozen, and often this leads to alternative splicing, which produces more than a single transcript, each of which is translated to produce more than one protein. For example, the dystrophin (mRNA) gene contains 78 spliceosomal introns, comprising more than 99% of the primary transcript (Tennyson et al. 1995). This gene encodes for several proteins, due to alternative splicing from the primary transcript.
Phylogenetically, group I introns have the widest distribution of all intron classes (Fig. 2), being present in the genomes of Archaea and Bacteria, as well as in the nuclear and organellar genomes of most phyla of Eukarya, and appear to be one of the most ancient classes of introns (Figs. 2, 3, 4). Group I introns also are numerous in the bacteriophage of ancient gram positive (monoderm) bacteria, and are sporadically found in the bacteriophage of more modern gram negative bacteria, again suggesting an ancient origin (Edgell et al. 2000). Archaeal introns have been found mainly in the tRNA and rRNA genes of Archaea and Eukarya, although remnants of the archaeal splicing mechanism are also found in rRNA processing, indicating a possible common ancient origin for archaeal introns and internal transcribed spacers in rRNA genes (Kjems and Garrett 1991; Belfort and Weiner 1997; Lykke-Anderson et al. 1997; Takai and Horikoshi 1999; Tang et al. 2002; Fujishima and Kanai 2014). The rRNA internal transcribed spacers in Bacteria and Archaea, and the rRNA ITS1 (internal transcribed spacer 1) in Eukarya may be the descendants of archaeal introns, and therefore they may have existed in the progenotes that preceded Bacteria. This would make archaeal introns, or their descendants, universal within all organisms, and suggests a mechanism for the joining of the SSU and LSU rRNA genes, which were separate during their early evolution (Fig. 1).
Group II introns have been reported in rRNA, tRNA, and mRNA genes of the organelles of opisthokonts, plants, and protists, as well as in the mRNA genes of Bacteria and Archaea. Spliceosomal introns only occur in the nuclear genomes of Eukarya, and were probably derived from a group II intron ancestor early in the evolution of Eukarya (at least 2.4 billion years ago; Rogozin et al. 2012). A reanalysis of an intron found in at least two genera of fungi (Phialophora and Cenococcum; Rogers et al. 1993; Shinohara et al. 1996) indicates that a group I and a group II intron exist at the same location within the same rDNA repeat near the end of the nuclear SSU rRNA (Fig. 5). This suggests a possible common origin for group I and group II introns. Also, it is possible that loci such as these provided opportunities to derive spliceosomal introns from group II introns.
Intron structures and evolution
Ribosomal RNA gene loci appear to be preferred targets for intron insertion, possibly because they are large loci, often occur in multiple copies, undergo frequent recombination events, and have high rates of transcription, potentially exposing them to intron insertion. In general, the possible routes of evolution for each type of intron (group I, group II, spliceosomal, and archaeal) can be deduced by their modes of splicing, modes of insertion, cell location, phylogenetic distribution, and genome locations (Fig. 2). As indicated above, group I introns have been reported in archaea, bacteria, the nuclear and organellar genomes of eukaryotes, and bacteriophage, while group II introns have been found in bacterial, archaeal, and eukaryotic organellar genomes (e.g., Bhattacharya et al. 2000, 2003; Perotto et al. 2000; Cannone et al. 2002; Jackson et al. 2002; Haugen et al. 2005; Simon et al. 2008; Chen 2010; Salman et al. 2012; Hausner et al. 2014). Group I and II introns splice via two transesterification reactions. While many can splice in vitro without the aid of other molecules, in vivo they rely on proteins to stabilize their catalytic conformations (Belfort et al. 1995; Zhang et al. 1995; Hausner et al. 2014). This is analogous to structural aspects of ribosomes, spliceosomes, and other cellular components based on associations between RNAs and proteins. For group I introns the first reaction is initiated by a free guanosine (GMP, GDP, or GTP, which associates with the P7 region) that becomes covalently bound to the 5′ end of the severed intron, while for group II introns, the first reaction is initiated by an internal adenine (within domain D6) that becomes covalently bound via its 2′ carbon to the 5′ end of the intron, becoming a lariat structure (Fig. 5). The second reactions are similar for both introns, which leads to complete release of the introns and fusing of the 5′ and 3′ exons in a standard 3′-5′ phosphodiester linkage.
Spliceosomal introns are likely reductions of group II introns, because their splicing reactions are identical to those of group II introns, although their lengths range from tens of nucleotides to hundreds of thousands of nucleotides. However, spliceosomal introns are not self-splicing, but require spliceosomes, which are composed of specific RNAs and proteins that bind at the exon–intron borders to affect splicing. Some of the proteins resemble ribosomal proteins, and some of the RNAs are similar to rRNAs (Kjems and Garrett 1991; Bleichert et al. 2006; Staley and Wolford 2009; Tocchini-Valentini et al. 2011). Thus, it is probable that they originated from some of the ribosomal proteins and rRNAs. Because of the connections between the introns, spliceosomal RNAs, rRNAs, ribosomal proteins, and spliceosomal proteins, it is evident that rRNAs and introns have been linked together, at least during portions of their evolutionary histories. However, spliceosomal introns have been found only in the nuclear genomes of eukaryotes (Fig. 2), which suggests a later origin (2.2–2.4 billion years ago), and a more limited distribution, for these introns. Although they are present in some of the ancient sections of the rRNAs, these may have been conversions from group II introns, or they may have been inserted later in rRNA evolution. Because they have not been found in organellar genomes in any eukaryote, they probably were present in the nuclear genomes of the first eukaryotes, and thus were not transferred to the nuclear genome from an organellar genome (the most common mode of movement of DNA within eukaryotic cells; Martin and Schnarrenberger 1997; Martin and Hermann 1998; Hackett et al. 2005; Rogers 2017).
Although archaeal introns appear to have had an evolutionary pathway independent of the other introns, some have proposed that they represent a lineage derived from a group I intron (Tochini-Valenti 2011; Nawrocki et al. 2018). Archaeal introns are present in some tRNA genes in species of Archaea, and they have also been found in nuclear tRNA and rRNA genes in a wide variety of eukaryotes (Kjems and Garrett 1991; Belfort and Weiner 1997; Lykke-Anderson et al. 1997; Bujnicki and Rychlewski 2001; Rodin and Rodin 2008; Fujishima et al. 2010; Rogers 2017; Fujishima and Kanai 2014). Splicing of these introns involves an endoribonuclease that recognizes a short bulge-helix-bulge (B-H-B) region that is formed by sequences of the 5′ and 3′ intron–exon borders that pair with one another. Interestingly, processing of the large rRNA precursor into the SSU rRNA and LSU rRNA in Archaea is nearly identical to the steps involved in cutting and joining the two exons away from the intron during splicing of archaeal introns (Belfort and Weiner 1997; Tang et al. 2002; Tocchini-Valentini et al. 2011). Therefore, parts of the rDNA internal transcribed spacer (ITS) regions in Archaea, Bacteria, and ITS1 in Eukarya may have originated from archaeal introns, which might initially have served to join the ancestral SSU and LSU rRNA genes together. In this case, intron splicing and parts of rRNA processing may have a common origin, suggesting the importance of archaeal introns in the evolution of ribosomal RNAs. In eukaryotes, ITS2 is an intervening sequence between the LSU rRNA and the 5.8S rRNA, but has had a different evolutionary pathway than ITS1, and has a different three dimensional structure (Edger et al. 2014; Giudicelli et al. 2014; Rogers, unpubished). The 5.8S gene and the 5′ end of the LSU rRNA pair in the mature ribosome, just as homologous regions pair in the LSU rRNAs of Bacteria and Archaea. However, bacterial and archaeal rRNAs are devoid of an ITS2 region, with the two sections of the LSU being contiguous. It is therefore possible that ITS2 originated as the insertion of an intron (with similarities to a group I intron) approximately 150 bp from the 5′ end of the LSU rRNA gene early in eukaryote evolution, separating the 5.8S rRNA gene from the remainder of the LSU rRNA gene.
Conversion among Introns
Introns of all four classes have been found inserted into neighboring nucleotide positions (Fig. 3) within the LSU rRNA (nucleotide positions 775–787, 858, 1024–1025, 1091–1098, 2076–2069, 2437–2455) and SSU rRNA (nucleotide positions 263–265, 287–300, 322–337, 788–793, 901–911, 939–943, 1057–1068, 1197–1201, 1224–1229, 1506–1521). In many cases, these introns exist either adjacent to, or overlapped with, other introns from different classes. In the case of some group I introns, there have been shifts in the splice site, yielding very similar introns with slightly different splice sites (Rogers et al. 1993; Gargas et al. 1995; Bhattacharya et al. 2000; Harris and Rogers 2008). The best example of this is near the 3′ end of the SSU gene in fungi (Figs. 3, 5, 7). Group I introns have been reported at nucleotide positions 1495, 1506, 1512, 1516 and 1521 (relative to those in the E. coli SSU gene), as well as at least two spliceosomal (or group II) introns at 1504 and 1512 (Rogers et al. 1993; Gargas et al. 1995; Bhattacharya et al. 2000; Jackson et al. 2002; Harris 2007; Harris and Rogers 2008, 2011). The concurrence of a spliceosomal/group II and a group I intron at the same location first was reported within the rRNA SSU genes of Phialophora americana and Cenococcum geophilum (Rogers et al. 1993; Yan et al. 1995; Shinohara et al. 1996; Harris 2007; Harris and Rogers 2008, 2011, 2014; Figs. 5, 7), two dematiaceous anamorphic fungal species that live in soils, as well as on plants and animals. This finding was initially surprising, because group II and spliceosomal introns were not known to occur in rRNA genes at that time, and the close proximities of group I, group II, and spliceosomal introns was also unknown. However, since then, many additional group I, group II, and spliceosomal intron positions have been catalogued, and some of them overlap with one another (Rogers et al. 1993; Bhattacharya et al. 2000; Cannone et al. 2002; Jackson et al. 2002; Harris and Rogers 2008; Figs. 3, 5, 7). This has provided support for the hypothesis that group I, group II, and spliceosomal introns may have evolved together within the rDNA loci. Folding into both group I and group II conformations is possible for some introns in this region, with only a slight shift of a few nucleotides in the putative splice sites (Fig. 5). Because of the sequence similarities in introns at the same positions, there were probably single insertions of group I or group II introns at several sites, followed by mutation and/or altered splicing of the introns, possibly aided by structures within the rRNA. This still leaves open the question of how some of these transitioned from group I introns into spliceosomal/group II introns, as well as whether they can be spliced by either of the two mechanisms under different conditions. Some ribosomal proteins have similarities to spliceosomal proteins (Fabrizio et al. 1997; Tang et al. 2002), and therefore, during splicing and processing of the rRNA, and assembly of the ribosomes in the nucleolus, some of the ribosomal proteins may have aided the introns to fold into a structure that could splice via a group II or spliceosomal splicing pathway, thus converting the group I intron into a group II or spliceosomal-type intron.
One set of short degenerate group I introns (62–78 nucleotides) located within the the SSU rRNA genes of several species of fungi (within the genera Arthonia, Phialophora, and Porpidia) exhibits yet another aspect of intron evolution within rRNA genes. These introns lack the central core regions (P2 through P8) of standard group I introns (Cech 1988, 1990; Cech and Herschlag 1996; Grube et al. 1996; Adams et al. 2004; Golden et al. 2005; Harris 2007; Harris and Rogers 2008, 2011, 2014; Fig. 7). When cloned and expressed as RNA, only the construct that included the short intron and the 3′ adjoining regions (containing the distal 3′ end of the SSU and the entire ITS1) exhibited group I intron self-splicing activity in vitro (Harris 2007; Harris and Rogers 2008, 2011, 2014; Fig. 6). Group I splicing activity could not be complemented by another functional group I intron that contained the core regions. Additionally, other recombinant constructs that lacked the 3′ regions (including ITS1), but included the small intron and sections of the 5′ regions (i.e., portions of the SSU rRNA), all failed to splice in vitro. This indicated that other portions of the RNA molecule, including ITS1, were capable of folding into a structure that effectively substituted for parts of an intron that had lost portions of its native structure, pointing to the malleable nature of rRNA and the rRNA introns. Interestingly, the remaining P9 region in the short introns appears to serve as the guanosine binding site, and contained a region that resembled the P7 guanosine binding site found in standard group I introns (Figs. 5, 6). This demonstrates the cross-functionality of sections of rRNAs and introns. ITS1 is also important in rRNA processing, which resembles archaeal intron splicing. Therefore, it is possible that parts of its structure have evolved to maintain conformations conducive to functions necessary for processing and/or splicing. As previously outlined, ITS1 appears to have similarities, and possible evolutionary linkage, with archaeal introns (Tang et al. 2002; Tocchini-Valentini et al. 2011), and therefore may maintain structures to assure functional conditions for splicing and/or rRNA processing.
Ribosomal loci as intron nurseries
Once introns contained genes for reverse transcriptases, riboendonucleases, endonucleases, transposases, and/or resolvases, they became mobile and could insert into RNA genes in an RNA/protein world, or into DNA genes within the DNA/RNA/protein world (Fig. 8). The fact that some of the archaeal intron endoribonucleases also exhibit endonuclease activity indicates that the transition from single stranded RNA to double stranded RNA and DNA templates might have occurred early in rRNA/rDNA evolution (Takai and Horikoshi 1999; Bujkicki and Rychlewski 2001; Tang et al. 2002; Fujishima et al. 2010). While the introns may have aided in the rapid evolution of rRNA and ribosomes, early organisms probably had only one rRNA gene (containing one copy each of the SSU and LSU sequences), thus limiting their rates of evolution and spread. Contemporary Bacteria possess from 1 to 15 rRNA gene loci per genome, and Archaea have 1–4 rRNA gene loci, while Eukarya have from one to tens of thousands of rDNA copies, and those that have only one copy have mechanisms for greatly increasing the rDNA copy numbers (Rogers and Bendich 1987a, b; Alberts et al. 1998; Rogers 2017). Organisms with multiple rRNA gene copies are better able to respond to changes in environmental conditions than are those with low rDNA copy numbers (Stevenson and Schmidt 2004), thus they can survive a larger number of evolutionary challenges than organisms with only a single or a few rRNA gene copies. Part of reason for this might be due to the dependence on the number of rDNA copies needed to produce the large number of rRNAs and ribosomes (approximately 20,000 to 60,000 per bacterial cell, and approximately 2 million to 10 million per eukaryotic cell). Cell cycle duration is also slower in organisms with low rDNA copy numbers, indicating that the number of rRNA genes may constitute significant controls on cell populations and developmental processes (Shermoen and Keifer 1975; Tartof 1975; Rogers 2017).
Having more rDNA copies implies that, frequently, there are more copies of the rRNA introns for those species with introns. The probabilities of intron mutation, transposition, and gain (and losses) are generally higher in species with multiple rRNA gene copies, and therefore, the rDNA loci could serve as evolutionary testing grounds for mutations, gains, and losses of these introns (Fig. 8). This is more exaggerated in eukaryotes, most of which have multiple tandem copies of rDNA. For example, arthropods usually have 100 or more rDNA tandem copies per haploid genome, mammals possess a few hundred tandem copies, and amphibians and plants have widely variable rDNA copy numbers from a few hundred to tens of thousands (Rogers et al. 1986; Rogers and Bendich 1987a, b; Rogers 2017). Introns associated with rDNA loci may have been able to spread and evolve rapidly, partly because of the relatively large numbers of rRNA gene repeats, intron copy number, frequent crossovers, gene conversion events, and availability of homing endonucleases or reverse transcriptases (Figs. 2, 8; Rogers and Bendich 1987a, b; Rogers 2017).
The presence of all four classes of introns in rDNA loci, and the reports of overlaps of different classes of introns, lend support to the hypothesis that rDNA loci have functioned as intron nurseries for hundreds of millions to billions of years (Jackson et al. 2002; Rogers et al. 1993; Gargas et al. 1995; Shivji et al. 1995; Bhattacharya et al. 2000, 2001; Itoh et al. 1998; Edgell et al. 2000; Roy et al. 2001, 2002; Haugen et al. 2005; Moriera et al. 2012; Figs. 2, 8). The rDNA loci may also have provided the raw materials for the evolution of spliceosomal introns, which are likely derivatives of group II introns. Bolstering this point, spliceosomes contain some proteins that are related to ribosomal proteins, which may place their likely origin close to the locations of ribosome assembly (i.e., the eukaryotic nucleolus) (Bleichert et al. 2006; Staley and Wolford 2009; Tocchini-Valentini et al. 2011). In addition, domain V of the LSU rRNA has similarities to spliceosomal U6/U2 RNA, again indicating close and ancient relationships between introns and rRNA (Kjems and Garrett 1991). While introns appear to have been intimately involved in the evolution of rRNAs, concurrently the rDNA loci may have provided opportunities for the diversification of introns, due to the functional plasticity of the RNAs, the multiple copies of the rRNA genes, the high rates of transcription, and the frequent recombination and gene conversion events within the rRNA gene locus. The overall conclusion is that rRNAs and introns collaborated from very early in rRNA evolution, which led both to a fully functional and complex ribosome, and to a diverse set of introns, both of which have been absolutely vital to all life on Earth.
References
Adams PL, Stahley MR, Gill ML, Kosek AB, Wang J, Strobel SA (2004) Crystal structure of a group I intron splicing intermediate. RNA 10:1867–1887
Alberts B, Bray D, Johnson A, Lewis J, Raff M, Roberts K, Walter P (1998) Essential cell biology. Garland, New York
Bada JL (2013) New insights into prebiotic chemistry from Stanley Miller’s discharge experiments. Chem Soc Rev 42:2186–2196
Baldauf SL (2003) The deep root of eukaryotes. Science 300:263–273
Belfort M, Weiner A (1997) Another bridge between Kingdoms: tRNA splicing in Archaea and Eukaryotes. Cell 89:1003–1006
Belfort M, Reaban ME, Coetzee T, Dalgaard JZ (1995) Prokaryotic introns and inteins: a panoply of form and function. J Bacteriol 177:3897–3903
Belousoff MJ, Davidovich C, Zimmerman E, Caspi Y, Wekselman I, Rocenszajn L, Shapira T, Sade-Falk O, Taha L, Bashan A, Weiss MS, Yonath A (2010) Ancient machinery embedded in the contemporary ribosome. Biochem Soc Trans 38:422–427
Bhattacharya D, Damberger S, Surek B, Melkonian M (1996) Primary and secondary structure analysis of the rDNA group I intron of the Zygnematales (Charophyta). Curr Genet 29:282–286. https://doi.org/10.1371/journal.pone.0101341
Bhattacharya D, Lutzoni F, Reeb V, Simon D, Fernandez F (2000) Widespread occurrence of spliceosomal introns in the rDNA genes of Ascomycetes. Mol Biol Evol 17:1971–1984
Bhattacharya D, Cannone JJ, Gutell RR (2001) Group I intron lateral transfer between red and brown algal ribosomal RNA. Curr Genet 40:82–90
Bhattacharya D, Simon D, Huang J, Cannnone JJ, Gutell RR (2003) The exon context and distribution of Euascomycetes rRNA spliceosomal introns. BMC Evol Biol 3:7
Bleichert F, Granneman S, Osheim YN, Baserga AL (2006) The PINc domain protein Utp24, a putative nuclease, is required for the early cleavage steps in 18S rRNA maturation. Proc Natl Acad Sci USA 103:9464–9469
Bokov K, Steinberg SV (2009) A hierarchical model for the evolution of 23S ribosomal RNA. Nature 457:977–980
Bujnicki JM, Rychlewski L (2001) Unusual evolutionary history of the tRNA splicing endonuclease EndA: relationship to the LAGLIDADG and PD-(D/E)XK deoxyribonucleases. Prot Sci 10:656–660
Caetano-Anollés G, Caetano-Anollés D (2015) Computing the origin and evolution of the ribosome from its structure—uncovering processes of macromolecular accretion benefits synthetic biology. Comput Struct Biotechnol J 13:427–447
Caetano-Anollés G, Wang M, Caetano-Anollés D (2013) Structural phylogenomics retrodicts the origin of the genetic code and uncovers the evolutionary impact of protein flexibility. PLoS One. 10:1371/journal.pone.0072225
Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Mueller KM et al (2002) The comparative RNA web (CRW) site: an online database of comparative sequence and structural information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2
Cech TR (1988) Conserved sequences and structures of group I introns: Binding an active site for RNA catalysis—a review. Gene 73:259–271
Cech TR (1990) Self-splicing group I introns. Ann Rev Biochem 59:543–568
Cech TR, Herschlag D (1996) Group I ribozymes: Substrate recognition, catalytic strategies, and comparative mechanistic analysis. Nucleic Acids Mol Biol 10:1–18
Chen X (2010) Evolution of group I introns in the nuclear ribosomal RNA genes of Dothidiomycetes. Thesis, Bowling Green State University, Bowling Green, OH
Chen W, Lynn EG, Grau CR (1998) Characterization of a group I intron in the nuclear rDNA differentiating Phialophora gregata f. sp. adzukicola from P. gregata f. sp. sojae. Mycoscience 39:279–283
Darnell JE, Doolittle WF (1986) Speculation on the early course of evolution. Proc Natl Acad Sci USA 83:1271–1275
Darnell J, Lodish H, Baltimore D (1990) Molecular cell biology. W.H. Freeman and Company, New York
Davila-Aponte J, Huss V, Sogin M, Cech TR (1991) A self-splicing group I intron in the nuclear pre-rRNA of the green alga, Ankistrodesmus stipitatus. Nucleic Acids Res 19:4429–4436
DeWachter R, Neefs J, Gori A, Van dePeer Y (1992) The gene coding for small ribosomal subunit RNA in the basidiomycete Ustilago maydis contains a group I intron. Nucleic Acids Res 20:1251–1257
Edgell DR, Belfort M, Shub DA (2000) Barriers to intron promiscuity in Bacteria. J Bacteriol 182:5281–5289
Edgell DR, Chalamcharla VR, Belfort M (2011) Learning to live together: mutualism between self-splicing introns and their hosts. BMC Biol 9:22
Edger PP, Tang M, Bird KA, Mayfield DR, Conant G, Mummenhoff K, Koch MA, Pires JC (2014) Secondary structure analysis of the nuclear rRNA internal transcribed spacers and assessment of its phylogenetic utility across the Brassicaceae (mustards). PLoS ONE 9(7):e101341
Fabrizio P, Laggerbauer B, Lauber J, Lane WS, Lührmann R (1997) An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J 16:4092–4106
Fox GE (2010) Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol 2(9):a003483
Fujishima K, Kanai A (2014) tRNA gene diversity in the three domains of life. Front Genet 5:1–11
Fujishima K, Sugahara J, Tomita M, Kanai A (2010) Large-scale tRNA intron transposition in the archaeal Order Thermoproteales represents a novel mechanism of intron gain. Mol Biol Evol 27:2233–2243
Gargas A, DePriest PT, Taylor JW (1995) Positions of multiple insertions in SSU rDNA of lichen-forming fungi. Mol Biol Evol 12:208–218
Gast RJ, Fuerst PA, Byers TJ (1994) Discovery of group I introns in the nuclear small subunit ribosomal RNA genes of Acanthamoeba. Nucleic Acids Res 22:592–596
Gilbert W, de Souza SJ, Long M (1997) Origin of genes. Proc Natl Acad Sci USA 94:7698–7703
Golden BL, Kim H, Chase E (2005) Crystal structure of an active group I ribozyme-product complex. Nat Struc Mol Bio 12:82–89
Grube M, Gargas A, DePriest PT (1996) A small insertion in the SSU rDNA of the lichen fungus Arthronia lapidicola is a degenerate group-I intron. Curr Genet 29:582–586
Guidicelli GC, Mäder G, Silva-Arias GA, Zamberlan PM, Bonatto SL, Freitas LB (2017) Secondary structure of nrDNA internal transcribed spacers as a useful tool to align highly divergent species in phylogenetic studies. Genet Molec Biol 40:191–199
Gutierrez G, Blanco O, Divakar PK, Lumbsch HT, Crespo A (2007) Patterns of group I intron presence in nuclear SSU rDNA of the lichen family Parmeliaeae. J Mol Evol 64:181–195
Hackett JD, Anderson DM, Erdner DL, Bhattacharya D (2004) Dinoflagellates: a remarkable evolutionary experiment. Am J Bot 91:1523–1534
Harish A, Caetano-Anollés G (2012) Ribosomal history reveals origins of modern protein synthesis. PloS ONE 7:e32776
Harris LB (2007) Characterization of a small ribozyme with self-splicing activity. Dissertation, Department of Biological Sciences, Bowling Green State University, Bowling Green, OH
Harris L, Rogers SO (2008) Splicing by an unusually small group I ribozyme. Curr Genet 54:213–222
Harris LB, Rogers SO (2011) Evolution of small putative group I introns in the SSU rRNA gene locus of Phialophora species. BMC Res Notes 4:258–262
Harris LB, Rogers SO (2014) Finding and characterizing small group I introns. In: Genomics III: methods, techniques, and applications. iConcept Press, Hong Kong, pp 327–341
Haugen P, Runge HJ, Bhattacharya D (2004) Long-term evolution of the S788 fungal nuclear small subunit rRNA group I introns. RNA 10:1084–1096
Haugen P, Simon D, Bhattacharya D (2005) The natural history of group I introns. Trends Genet 21:111–119
Hausner G, Hafez M, Edgell DR (2014) Bacterial group I introns: mobile RNA catalysts. Mob DNA 5:8
Huang L, Krupkin M, Bashan A, Yonath A, Massa L (2013) Protoribosome by quantum kernel energy method. Proc Natl Acad Sci USA 110:14900–14905
Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Jillian F (2016) A new view of the tree of life. Nature Microbiol 1:16048. https://doi.org/10.1038/nmicrobiol.2016.48
Itoh T, Suzuki K, Nakase T (1998) Occurrence of introns in the 16S rRNA genes of members of the genus Thermoproteus. Arch Microbiol 170:155–161
Iwasa J, Marshall W (2016) Karp’s cell and molecular biology, 8th edn. Wiley, Hoboken
Jackson SA, Cannone JJ, Lee JC, Gutell RR, Woodson SA (2002) Distribution of rRNA introns in the three-dimensional structure of the ribosome. J Mol Biol 323:35–52
Johansen S, Vogt VM (1994) An intron in the nuclear ribosomal DNA of Didymium iridis codes for a group I ribozyme and a novel ribozyme that cooperates in self-splicing. Cell 76:725–734
Kanai A (2015) Disrupted tRNA genes and tRNA fragments: a perspective of tRNA evolution. Life 5:321–331
Kjems J, Garrett RA (1991) Ribosomal RNA introns in archaea and evidence for RNA conformational changes associated with splicing. Proc Natl Acad Sci USA 88:439–443
Lickey EB, Hughes KW, Petersen RH (2003) Variability and phylogenetic incongruence of an SSU nrDNA group I intron in Artomyces, Aurscalpium, and Lentinellus (Auriscalpiaceae: Homobasidiomycetes). Mol Biol Evol 20:1909–1916
Lykke-Anderson J, Aagaard C, Semionenkov M, Garrett RA (1997) Archaeal introns: splicing, intercellular mobility and evolution. TIBS 22:326–331
Martin W, Hermann RG (1998) Gene transfers from the organelles to the nucleus: how much, what happens, and why? Plant Phys 118:9–17
Martin W, Schnarrenberger C (1997) The evolution of the Calvin cycle from prokaryotic to eukaryotic chromosomes: a case study of functional redundancy in ancient pathways through endosymbiosis. Curr Genet 32:1–18
Michel F, Westhof E (1990) Modeling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol 216:585–610
Michel F, Umesono K, Ozeki H (1989) Comparative and functional anatomy of group II catalytic introns—a review. Gene 82:5–30
Moreira S, Breton S, Burger G (2012) Unscrambling genetic information at the RNA level. WIREs RNA. https://doi.org/10.1002/wrna.1106
Muller KM, Cannone JJ, Gutell RR, Sheath RG (2001) A structural and phylogenetic analysis of the group IC1 introns in the order Bangiales (Rhodophyta). Mol Biol Evol 18:1654–1667
Murphy FL, Cech TR (1994) GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J Mol Biol 236:49–63
Nawrocki EP, Jones TA, Eddy SR (2018) Group I introns are widespread in archaea. Nucleic Acids Res 46:7970–7976
Nikoh N, Fukatsu T (2001) Evolutionary dynamics of multiple group I introns in nuclear ribosomal RNA genes of endoparasitic fungi of the genus Cordyceps. Mol Biol Evol 18:1631–1642
Perotto S, Nepote-Fus P, Saletta L, Bandi C, Young JPW (2000) A diverse population of introns in the nuclear ribosomal genes of ericoid mychorrhizal fungi includes elements with sequence similarity to endonuclease-coding genes. Mol Biol Evol 17:44–59
Petrov AS, Bernier CR, Hsaio C, Norris AM, Kovacs NA, Waterbury CC, Stepanov VG, Harvey SC, Fox G, Wartell RM, Hud NV, Williams LD (2014) Evolution of the ribosome at atomic resolution. Proc Natl Acad Sci USA 111:10251–10256
Petrov AS, Gulen B, Norris AM, Kovacs NA, Bernier CR, Lanier KA, Fox GE, Harvey SC, Wartell RM, Hud NV, Williams LD (2015) History of the ribosome and the origin of translation. Proc Natl Acad Sci USA 112:15396–15401
Roberts E, Eargle J, Wright D, O’Donoghue P, Sethi A, Dhaliwal B (2008) Evolutio of biomolecular structure. Luthey-Schulten group. University of Illinois, Urbana-Champaign
Roberts E, Eargle J, Wright D, Li L, Sethi A, Luthey-Schulten Z (2011) Evolution of translation. Luthey-Schulten group. University of Illinois, Urbana
Rodin SN, Rodin AS (2008) On the origin of the genetic code: signatures of its primordial complementarity in tRNA and aminoacyl-tRNA synthetases. Heredity 100:341–355
Rodin AS, Szathmáry E, Rodin SN (2011) On origin of genetic code and tRNA before translation. Biol Direct 6:14
Roger AJ, Simpson AGB (2009) Evolution: revisiting the root of the eukaryote tree. Curr Biol 19:R165–R167
Rogers SO (2017) Integrated molecular evolution, second edn. CRC Press, Taylor & Francis Group, Boca Raton
Rogers SO, Bendich AJ (1987a) Heritability and variability in ribosomal RNA genes of Vicia faba. Genetics 117:285–295
Rogers SO, Bendich AJ (1987b) Ribosomal RNA genes in plants: variability in copy number and in the intergenic spacer. Plant Mol Biol 9:509–520
Rogers SO, Bendich AJ (1988) Recombination in E. coli between cloned ribosomal RNA intergenic spacers from Vicia faba: a model for the generation of ribosomal RNA gene heterogeneity in plants. Plant Sci 55:27–31
Rogers SO, Honda S, Bendich AJ (1986) Variation in ribosomal RNA genes among individuals of Vicia faba. Plant Mol Biol 6:339–345
Rogers SO, Yan ZH, LoBuglio KF, Shinohara M, Wang CJK (1993) Messenger RNA intron in the nuclear 18S ribosomal DNA gene of deuteromycetes. Curr Genet 23:338–342
Rogozin IB, Carmel L, Csuros M, Koonin EV (2012) Origin and evolution of spliceosomal introns. Biol Direct 7:11
Root-Bernstein M, Root-Bernstein R (2015) The ribosome as a missing link in the evolution of life. Theoret Biol 367:130–158
Roy SW, Lewis BP, Fedorov A, Gilbert W (2001) Footprints of introns on the eukaryotic genome. Trends Genet 17:496–499
Roy SW, Fedorov A, Gilbert W (2002) The signal of ancient introns is obscured by intron density and homolog number. Proc Natl Acad Sci USA 99:15513–15517
Salman V, Amann R, Shub DA, Shulz-Vogt HN (2012) Multiple self-splicing introns in the 16S rRNA genes of giant sulfur bacteria. Proc Natl Acad Sci 109:4203–4208
Seetharaman M, Eldho NV, Padgett RA, Dayie KT (2006) Structure o a self-splicing group II intron catalytic effector domain 5: paralleles with spliceosomal U6 RNA. RNA 12:235–247
Shermoen AW, Keifer BI (1975) Regulation in rDNA-deficient Drosophila melanogaster. Cell 4:275–280
Shinohara ML, LoBuglio KF, Rogers SO (1996) Group-I intron family in the nuclear ribosomal RNA small subunit genes of Cenococcum geophilum. Curr Genet 29:377–387
Shivji M, Rogers SO, Stanhope M (1995) Molecular studies on sharks. Shark Tagger 1995 Summary, NOAA. p 15
Simon DM, Clarke NAC, McNeil B, Johnson I, Pantuso D, Dai L, Chai D, Zimmerly S (2008) Group II introns in Bacteria and Archaea: ORF-less introns and new varieties. RNA 14:1704–1713
Staley JP, Wolford JL Jr (2009) Assembly of ribosomes and spliceosomes: complex ribonuclear machines. Curr Opin Cell Biol 21:108–118
Stevenson BS, Schmidt TM (2004) Life history implications of rRNA gene copy number in Escherichia coli. Appl Environ Microbiol 70:6670–6677
Takai K, Horikoshi K (1999) Molecular phylogenetic analysis of Archaeal intron-containing genes coding for rRNA obtained from a deep-subsurface geothermal water pool. Appl Environ Microbiol 65:5586–5589
Tang TH, Rozhdestvensky TS, d’Orval BC, Bortolin M-L, Huber H, Charpentier B, Branlant C, Bachellerie J-P, Brosius J, Hüttenhofer A (2002) RNAomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res 30:921–930
Tartof KD (1975) Redundant genes. Annu Rev Genet 9:355–385
Tennyson CN, Klamut HJ, Worton RG (1995) The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nature Genet 9:184–190
Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP (2011) Evolution of introns in the archaeal world. Proc Natl Acad Sci USA 108:4782–4787
Wekselman I, Davidovich C, Agmon I, Zimmerman E, Rozenberg H, Bashan A, Berisio R, Yonath A (2009) Ribosome’s mode of functions: myths, facts and recent results. J Pept Sci 15:122–130
Yan ZH, Rogers SO, Wang CJK (1995) Assessment of Phialophora species based on ribosomal DNA internal transcribed spacers and morphology. Mycologia 87:72–83
Zuker M (2003) Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:1–10
Acknowledgements
I thank students in my Integrated Molecular Evolution course who helped me to hone some of the ideas and explanations presented here. I thank Lorena Harris, Mari Shinohara, Zhonghua Yan, Xing Chen, Kathy LoBuglio, CJK Wang, Mahmood Shvji, and Mike Stanhope who participated in various projects associated with the research presented in this study.
Author information
Authors and Affiliations
Contributions
SOR conceived and wrote all portions of the paper, and drew all of the diagrams.
Corresponding author
Ethics declarations
Conflict of interest
The author (SOR) declares that he has no conflicts of interest.
Ethical approval
No human subjects were used in this study. Only fungal and bacterial cultures were used.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.