Abstract
Of the six known autotrophic pathways, the Wood-Ljungdahl pathway (WL) is the only one present in both the acetate producing Bacteria (homoacetogens) and the methane producing Archaea (hydrogenotrophic methanogens), and it has been suggested that WL is one of the oldest metabolic pathways. However, only the so-called carbonyl branch is shared by Archaea and Bacteria, while the methyl branch is different, both in the number of reactions and enzymes, which are not homologous among them. In this work we show that some parts of the methyl branch of archaeal Wood-Ljungdahl pathway (MBWL) are present in bacteria as well as in non-methanogen archaea, although the tangled evolutionary history of MBWL cannot be traced back to the Last Common Ancestor. We have also analyzed the different variants of methanogenesis (hydrogenotrophic, acetoclastic and methylotrophic pathways), and concluded that each of these pathways, and every different enzyme or subunit (in the case of multimeric enzymes), has their own intricate evolutionary history. Our study supports the scenario of hydrogenotrophic methanogenesis being older than the other variants, albeit not old enough to be present in the last archaeal common ancestor.
Similar content being viewed by others
Introduction
The molecular details of universally distributed biological processes not only suggest the monophyletic origin of all extant forms of life, but also imply that the sets of genes encoding these complex traits became fixed long time ago. All organisms share the same genetic code, the same essential features of genome replication and gene expression, basic anabolic reactions, and membrane-associated ATPase-mediated energy production, along with other basic abilities, derived from a common ancestral form, i.e., the last common ancestor (LCA, a.k.a. LUCA). All known biological variations can be easily understood as the outcome of divergent processes from an ancestral life form that predated the separation of the Archaea and Bacteria domains (Becerra et al. 2007).
During the past two decades, different attempts have been made to describe the nature of the LCA (Becerra et al. 2007). The results obtained include lists of repertoires of gene sequences from incompletely represented basic biological processes, such as transcription, translation, energy metabolism, biosyntheses of nucleotides and amino acids, as well as some sequences related to replication, repair, and cellular transport. In spite of the limitations of the different methodological approaches employed, these inventories provide significant insights into LCA biological traits. However, these reconstructions are less informative about the metabolic abilities of the LCA. Some authors propose that together with acetogenesis, methanogenesis is one of the oldest metabolisms on Earth (Martin and Russell 2007; Sousa et al. 2013), and suggest that both metabolic routes emerged in a hyperthermophilic environment, such as the alkaline hydrothermal environment (Martin and Russell 2007), from an LCA endowed with geochemically-driven monocarbon-unit (C1) transformations (Sousa and Martin 2014; Weiss et al. 2016). The available information suggests that methanogenesis is present only in the Archaea domain, whereas the acetyl-CoA synthesis from CO2, or Wood-Ljungdahl (WL) pathway, is present in both Bacteria and Archaea. Among the six known autotrophic routes, plus the recent discovery of a canonical TCA cycle flowing in reverse (Mall et al. 2018; Nunoura et al. 2018), the WL pathway is the only one that may conserve energy in addition to fixing carbon, it is a relatively simple linear route and not an autocatalytic cycle (Hügler and Sievert 2011; Peretó 2012). These traits have led to the proposal that it is the oldest autotrophic metabolism on Earth (Peretó et al. 1999; Berg et al. 2010; Fuchs 2011) that may have already been present in the LCA as a primitive, geochemically-driven version (Weiss et al. 2016).
Biologically, methane production is limited to some members of the Archaea under anaerobic conditions (Whitman et al. 2006) as well as to bacteria containing Fe-only nitrogenase (Zheng et al. 2018). Archaeal methanogenesis is the main source for biogenic methane and can follow three metabolic variants: (i) the hydrogenotrophic pathway based on CO2 and H2; (ii) the acetoclastic route; and (iii) those based on methyl compounds (methylotrophic) (Ferry 1999; Whitman et al. 2006; Costa and Leigh 2014). The most important source of methane on Earth is the split of acetate, which is responsible for two-thirds of the total amount produced during the last 300 years (Ferry 2010), whereas the reduction of CO2 and the use of methylated compounds have produced the remaining amount. Geological reports of methane inclusions of possible biological origin would indicate that methanogenesis is at least 3.4–3.5 Gyr old (Ueno et al. 2006). Moreover, molecular clock analyses calibrated with horizontal gene transfer events place the divergence of this metabolism within the phylum Euryarchaeota no later than 3.5 Gyr ago (Wolfe and Fournier 2018).
Over a decade ago, all known methanogenic Archaea were believed to be part of the Euryarchaeota phylum and could not be placed in a monophyletic group. This lead to the proposal that methanogens could be divided into two classes (Bapteste et al. 2005). It was argued that methanogenesis arose earlier within this phylum (Bapteste et al. 2005; Gribaldo and Brochier-Armanet 2006), was inherited vertically, and lost independently multiple times during the evolutionary history of Euryarchaeota (Borrel et al. 2013; Gao and Gupta 2007; Gribaldo and Brochier-Armanet 2006). Recently, the hypothesis that methanogenesis is restricted to the Euryarchaeota has been challenged by Evans et al. (2015), who suggested that the discovery of partial genome sequences isolated from environmental samples of a deep aquifer were part of the new non-Euryarchaeota phylum, designated as “Candidatus Bathyarchaeota”, that some members included the gene repertoire necessary for a methanogenesis based on methyl C1 compounds (Evans et al. 2015). Analysis of new metagenomic samples from anaerobic digesters with a high methane flux led to the identification of the new phylum “Ca. Verstraetearchaeota”, which together with the “Ca. Bathy”-, “Ca. Geo-”, Thaum-, Cren- and Korarchaeota, form the superphylum TACK or Proteoarchaeota (Spang et al. 2017). Members of “Ca. Verstraetearchaeota” are theoretically able to produce methane using methylated compounds, specifically methanol, methanethiol and methylamine (Vanwonterghem et al. 2016). Quite surprisingly, the findings of Laso-Pérez et al. (2016) suggest that different variants of the enzyme involved in the last step of methane biosynthesis (i.e. methyl-coenzyme M reductase or MCR) can oxidize butane in a reaction that resembles “reverse methanogenesis” in anaerobic methane oxidant (ANME) organisms. Finally, the characterization of two nearly completed sequenced genomes isolated from hypersaline lakes that belong to a deep-branching Haloarchaea, the Methanonatronarchaeia, a novel euryarchaeal group of extreme halophilic methyl-reducing methanogens using C1 compounds, expands our knowledge of organisms capable of performing methanogenesis within the Euryarchaeota phylum (Sorokin et al. 2017). All of these findings indicate a much more widespread presence of methanogenesis within the Archaea than previously suspected, suggesting the early origin of this metabolic mode in the archaeal domain’s ancestor, followed by multiple losses in many different lineages that are no longer capable of methane production (Borrel et al. 2016; Williams et al. 2017).
Here we examine the phylogenetic distribution and evolution of the enzymes and the coenzymes that are essential for the different methanogenic pathways, i.e., the hydrogenotrophic, acetoclastic, and the methylotrophic routes. Our results indicate that the early evolution of methanogenesis was subject of intense horizontal gene transfers (HGT). Moreover, the uneven phylogenetic distribution of the idiosyncratic methanogenic coenzymes also suggest that the last archaeal common ancestor was not a methanogen. As a consequence, the results presented here show that methanogenic pathways cannot be used as proxies of primordial life metabolic abilities, and even less as evidence of an autotrophic origin of life in hydrothermal environments rich in transition-metal sulfides.
Material and Methods
In this work, local databases were constructed with complete proteomes, which were retrieved from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al. 2014), Reference Sequence (RefSeq) database of National Center for Biotechnology Information (NCBI) (Pruitt et al. 2007) and Joint Genome Institute’s Integrated Microbial Genomes (JGI) (Markowitz et al. 2012) databases (December of 2016 for Bacteria and Eukarya) and December of 2017 for Archaea. For the “Ca. Verstraetearchaeota” genomes we followed the methodology of Vanwonterghem and collaborators using OrfM (Woodcroft et al. 2016) (https://github.com/wwood/ OrfM.git). An exhaustive homology search was performed for each one of the enzymes (and for each subunit of multimeric enzymes) of hydrogenotrophic, acetoclastic and methylotrophic methanogenesis pathways using BlastP search (Altschul et al. 1997), identity values of 35%, query coverage of 85% and e-value lower than 1 × 10−6 was taken as a cutoff. From BlastP search, sequences that pass the cutoff were aligned with MUSCLE v3.8.31 (Edgar 2004), using default parameters. An HMM profile was constructed with HMMER (Finn et al. 2011), and was used to scan the local databases, with the same values used in BlastP. Results of the HMMER search were aligned using MUSCLE. To improve the alignments, non-informative parts of the alignment (poorly aligned regions and big-gaps), were removed automatically using trimAL algorithm (Capella-Gutiérrez et al. 2009), with the “automated1” option. The substitution model selection for each alignment and inference of maximum likelihood trees was performed using IQ-TREE software (Nguyen et al. 2015), we obtained bootstrap supports values using an ultrafast bootstrap approximation (Minh et al. 2013) with 1000 replicates. Visualization, annotation and generation of images of each tree was made with iTOL software (Letunic and Bork 2016). A R script was made specifically to create the heatmaps in R v3.1.1 using phetamap and RcolorBrewer libraries. Phylogenetic trees for each gene based on untrimmed alignments were also performed (FASTA and Newick format files are available in figshare repository at https://figshare.com/s/6d27214f324ee631a586).
Results
Hydrogenotrophic Methanogenesis
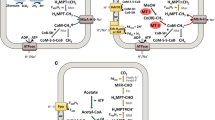
As shown in Fig. 1a, there are seven enzymatic steps involved in the reduction of CO2 to methane, in which several different coenzymes participate as C1- or electron-carriers (Ferry 1993; Graham and White 2002). The first enzyme that takes part in the reduction of CO2 to methane is the six subunits formylmethanofuran dehydrogenase (FmdA-F, EC 1.2.7.12), that reduces CO2 to formyl. There are two known isoenzymes, one containing molybdenum and another one that uses tungsten in the catalytic center. In this work, we have analyzed separately the different subunits of the molybdenum-containing enzyme (FmdA-F). Our results show a distinctive evolutionary path for each subunit, in which FmdA and FmdF are present in the three domains of life (Figs. S1 and S6), while FmdB, FmdC, FmdD and FmdE (Figs. S2-S5) are found only in Archaea and Bacteria. The enzymes catalyzing the second and third steps, formylmethanofuran tetrahydromethanopterin formyltransferase (Ftr, EC 2.3.1.101) (Fig. S7) and methenyl-H4MPT cyclohydrolase (Mch, EC 3.5.4.27) (Fig. S8), are single unit enzymes, and are present only in Bacteria and Archaea. The next step is carried out by two different enzymes, one H2-dependent, the N5, N10-methylene-tetrahydromethanopterin dehydrogenase (Hmd, EC 1.12.98.2) (Fig. S10), and another coenzyme-F420-dependent dehydrogenase (Mtd, EC 1.5.98.1) (Fig. S9) that participates in the so called Hmd-Mtd cycle. These enzymes are restricted to Archaea, except for one Desulfurobacterium thermolithotrophum (Bacteria), that is endowed with the H2-forming enzyme, the methylene-tetrahydromethanopterin dehydrogenase (Hmd) (Table S1 and Fig. S10). The methylene-H4MPT reductase (Mer, EC 1.5.98.2) (Fig. S11), reduces the methenyl group to methyl and is broadly distributed in both Bacteria and Archaea (Table S1). In the last steps the methyl group is finally transferred, first from H4MPT to factor III, and then to coenzyme M by the methyl-H4MPT coenzyme M methyltransferase (EC 2.1.1.86) complex, which is located in the cellular membrane, and is composed of eight subunits (MtrA, MtrB, MtrC, MtrD, MtrE, MtrF, MtrG and MtrH). This enzyme is mainly restricted to the Archaea domain, specifically the MtrB, MtrC, MtrD, MtrE, MtrF and MtrG subunits, which are confined to the Euryarchaeota phylum (Figs. S13-S18). On the other hand, MtrA (Fig. S12) is present in members of Euryarchaeota as well as in a few members of Thaumarchaeota, while MtrH (Fig. S19) has a wide distribution in Archaea and is found in “Ca. Loki-”, “Ca. Thor-”, “Ca. Odin-” “Ca. Bathy-”, “Ca. Verstraete-”, Cren- and Korarchaeota phyla, and in Bacteria in some members of Deltaproteobacteria, Alphaproteobacteria and Firmicutes-Clostridia.
Methabolic variants of biological production of methane, circles represent presence and absence of the different enzymes among the tree domains of life (Eukarya in red, Archaea in blue and Bacteria in green). a Hydrogenotrophic Methanogenesis in which Archaeal MBWL comprise the first five metabolic steps (from Fmd to Mer), Mtr are shared with Acetoclastic Methanogenesis and Mcr are shared with all organisms able to do methane and in fact with all metabolic pathways of production of methane. b Acetoclastic Methanogenesis in which the first two steps vary from Methanosaeta and Methanosarcina species (see details in text). c Methylotrophic Methanogenesis, this is the most variant type of methanogenesis in terms of initial substrates and methyl transferases. Fmd Formylmethanofuran dehydrogenase. Ftr Formylmethanofuran:H4MPT formyltransferase. Mch Methenyl:H4MPT cyclohydrolase. Mtd-Hmd H2-forming methylene-H4MPT dehydrogenase. Mer F420-dependent methylene-H4MPT reductase. Mtr Methyl-H4MPT:coenzyme M methyltransferase. Mcr Methyl-coenzyme M reductase. Ack Acetate Kinase. Pta Phosphotransacetylase. Cdh protein of Acetyl-CoA decarbonylase/synthase complex. MtaB and MtaA methanol specific methyltransferase, MtmB Monomethylamine specific methyltransferase. MtbB Dimethylamine specific methyltransferase. MttB Trimethylamine specific methyltransferase. MtbA methyltransferase shared between methilamines. MtsA Dimethylsulfide specific methyltransferase. MtpA Methylpropianate specific methyltransferase
In the last enzymatic step, the methyl group is reduced to methane. This reduction is catalyzed by methyl coenzyme M reductase (Mcr, EC 2.8.4.1). The three subunits of this enzyme (McrA, McrB and McrG) are restricted to the Archaea domain, specifically in the Eury-, “Ca. Bathy-” and “Ca. Verstraetearchaeota” phyla (Figs. S20-S22). The enzyme Mtr is shared among all acetoclastic and hydrogenotrophic methanogens, while Mcr catalyzes the last crucial step for all metabolic routes of oxidation of methane, butane and production of methane in organisms capable of carrying out methanogenesis, anaerobic oxidation of methane (AOM) (Timmers et al. 2017) and butane oxidation (Laso-Pérez et al. 2016).
Acetoclastic Methanogenesis
The only two known genera capable of performing acetoclastic methanogenesis are Methanosarcina and Methanosaeta (Ferry 1992, 1999; Smith and Ingram-Smith 2007). In Methanosarcina the combination of acetate kinase (Ack, EC 2.7.2.1) and phosphotransacetylase (Pta, EC 2.3.1.8) produces acetyl-CoA from acetate (Ferry 1992; Lessner 2009). On the other hand, in Methanosaeta these two coupled reactions are carried out by an AMP-forming acetyl-CoA synthetase (ACS, EC 6.2.1.1) (Jetten et al. 1989). The next reaction is catalyzed by CO dehydrogenase/acetyl-CoA synthase (CODH/ACS, EC 2.3.3.-) (Ferry 1997), which transfers the methyl group to tetrahydrosarcinopterin (H4SPT). A methyltransferase then transfers the methyl group from H4SPT to coenzyme M (CoM) and, finally, methyl-CoM reductase reduces the methyl group to methane (Ferry 1997) (see Fig. 1b).
Our results indicate that Ack (Fig. S23) is a generalist enzyme capable of activating-chemically acetate by ATP-dependent phosphorylation. It is broadly distributed in Bacteria, as well as in Eukarya (in fungi, specifically in Basidiomycetes and Ascomycetes, some plants, green algae, and protists-amebozoa), while in Archaea it is present in only Methanosarcina species as well as in “Ca. Pacearchaeota” and “Ca. Woesearchaeota”. On the other hand, Pta (Fig. S24) is widely distributed in Bacteria, while in Archaea it is only present in Methanosarcina, Methanobacterium and “Ca. Woesearchaeota”.
The CODH/ACS subunits that we have analyzed belong to class II CODH/ACS’s (Lindahl and Chang 2001), which is different of the previously analyzed class I CODH/ACS’s operating in WL (Becerra et al. 2014). Our results indicate that each of the CODH/ACS subunits has a different evolutionary history. The phylogenies suggest that CdhB (EC 1.2.7.4) (Fig. S26) is a subunit that has an archaeal distribution with one exception within the Bacteria “Ca. Desulforudis audaxviator”, which appear to be the result of horizontal gene transfers events. The subunits CdhA (EC 1.2.7.4) (Fig. S25), CdhC (EC 2.3.1.169) (Fig. S27), CdhD (EC 2.1.1.245) (Fig. S28) and CdhE (EC 2.1.1.245) (Fig. S29) are broadly distributed among Archaea and Bacteria, as reported by Adam et al. (2018).
Methyl Compound-Based Methanogenesis
The utilization of methyl compounds as precursors in methane synthesis is confined to a small group of methanogens that belong to the Methanosarcinales (Methanosarcina and Methanolobus species), Methanomassiliicoccales (“Ca. Methanomassiliicoccus”) and Methanobacteriales (Methanosphaera) species, which exploit methanol, methylamines and methylated thiols (Liu and Whitman 2008) and, perhaps also to some organisms belonging to the “Ca. Bathyarchaeota” and “Ca. Verstraetearchaeota” (Evans et al. 2015; Vanwonterghem et al. 2016). The enzyme responsible for the utilization of methanol is methanol-CoM methyltransferase, which has three subunits MtaA (EC 2.1.1.246), MtaB (EC 2.1.1.90) and the non-catalytic MtaC. Only MtaA and MtaB have methyltransferase activity. Our phylogenetic analysis indicates a broad distribution of MtaA (Fig. S30) in Eukarya, Bacteria and Archaea, which suggests an intricate evolutionary history that indicates that the gene has undergone multiple horizontal gene transfer events. On the other hand, MtaB (Fig. S31) is present in Archaea, specifically in members of Euryarchaeota, and as well as in some members of Bacteria belonging to the Firmicutes-Clostridia and Deltaproteobacteria.
In the case of methylamine-dependent methanogenesis, the substrates monomethylamine, dimethylamine and trimethylamine are used by two different enzymes with methyltransferase activity. The first one is specific and it changes depending of the substrate (see Fig. 1c), while the second one, the methylated methylamine-specific corrinoid protein CoM methyltransferase (MtbA, EC 2.1.1.247), catalyzes a key step after one of the three different methylamines products (MtmB, MtbB or MttB) (Fig. 1c). For monomethylamine, the first protein with methyltransferase activity is monomethylamine methyltransferase (MtmB, EC 2.1.1.248), which has a broad distribution in Archaea and is also found in some members of Bacteria that belong to Firmicutes. It was thought that this enzyme was restricted to Euryarchaeota and “Ca. Verstraetearchaeota”. However, it was found in Euryarchaeota, Crenarchaeota as well as in “Ca. Verstraetearchaeota” (Fig. S32). This enzyme probably first evolved in Archaea and the gene then underwent HGT to Bacteria. In the case of dimethylamine, the first enzyme with methyltransferase activity is dimethylamine methyltransferase (MtbB, EC 2.1.1.249), which is restricted to some members of the Archaea domain, including members of Methanosarcina, Methanolobus, “Ca. Methanomassiliicoccus” and “Ca. Verstraetearchaeota”, and only one Firmicutes-Clostridia (SF, S33) (Thermacetogenium phaeum), which is an acetate-oxidizing thermophilic bacterium. For trimethylamine, we studied the trimethylamine methyltransferase (MttB, EC 2.1.1.250). Our phylogenies indicate a very restricted distribution to some members of Archaea, all belonging to Euryarchaeota phylum, and few members of Bacteria belonging to Firmicutes-Clostridia (Fig. S34). Finally, the enzyme present in all pathways using methylamine compounds is MtbA (Fig. 1c). Phylogenetic analysis of this enzyme (Fig. S35) shows a wide distribution in Bacteria, while in Archaea is restricted to Eury-, “Ca. Micra-”, “Ca. Geo-”, “Ca. Heimdall-”, “Ca. Thor-”, “Ca. Verstraete-” and one member of “Ca. Lokiarchaeota” phyla (see Table S1). In Eukaryotes it is found in some members of fungi, plants, green algae, protists and animals (Fig. S35).
Dimethylsulfide protein methyltransferase (MtsA, EC 2.1.1.251) is used for methylated thiols (dimethylsulfide) and has a broad distribution among Bacteria, but in Archaea it is confined to Eury-, “Ca. Bathy-”, “Ca. Loki-”, “Ca. Micra-”, “Ca. Geo-”, “Ca. Heimdall-”, “Ca. Thor-”, “Ca. Verstraetearchaeota” and some unclassified Archaea. Moreover, it is present in protists, fungi, animals, green algae and plants (SF, S36). The analysis of the phylogenetic distribution of the 3-(methylthio) propanoate coenzyme M methyltransferase (MtpA, EC 2.1.1.251) that uses as substrate methylmercaptopropianate, shows that it is present in Bacteria, while in Archaea it is found in members of Eury-, “Ca. Heidall-”, “Ca. Thor-” and “Ca. Verstraetearchaeota” phyla (Fig. S37 and Table S1). Unlike other methylated compounds methyltransferases, MtpA supplies two enzymes with methyltransferase function and therefore catalyzes two different reactions instead of only one.
As shown in supplementary material S45, trees constructed from non-trimmed alignments do not provide better resolution or substantially different topologies.
Use of Coenzymes in Methanogenesis
It is well known that nearly half of the characterized enzymes require organic cofactors or metal ions to carry out specific chemical reactions and enhance their catalytic power (Fischer et al. 2010). Enzymes involved in methanogenesis use different specific coenzymes in each step of their methane producing routes (Lessner 2009). The factor F430, coenzyme M and coenzyme B play a key role in the last step of methane biosynthesis (Graham and White 2002). The absence of one of them interrupts this biological process. As shown in Fig. 3, organisms from Euryarchaeota phylum contain the full repertoire of enzymes required for the synthesis of F430, M and B coenzymes. Furthermore, other organisms from the Archaea and Bacteria domains (Fig. 3 and Fig. S38) have only some coenzyme B biosynthetic enzymes that partake in other metabolic functions, like the α-aminoadipic acid lysine biosynthesis (Drevland et al. 2008; Kobashi et al. 1999). Therefore, it is not surprising to find this group of enzymes in other living beings (e.g. Bacteria domain). It has been argued that since the sequences of the key Mcr enzyme and some enzymes of the methylotrophic pathway are present in some members of “Ca. Bathyarchaeota” and “Ca. Verstraetearchaeota” these organisms maybe have the ability to carry out methanogenesis based on methylated compounds (Evans et al. 2015; Vanwonterghem et al. 2016) or in the case of “Ca. Bathyarchaeota” a few authors propose that some members of this phylum can carry out acetogenesis with the battery of the archaean WL (He et al. 2016). However, our results show that in “Ca. Bathyarchaeota” the enzymes required to synthesize F430 are absent, as well as the majority of the enzymes for the biosynthesis of M coenzyme. On the other hand, “Ca. Verstraetearchaeota” lack the enzymes required for M coenzyme biosynthesis (Fig. 3). All methanogenic organisms have the enzymes required to synthesize F430 factor, while other non-methanogenic organisms, including bacteria, only have the first enzymes involved in this metabolic pathway. This distribution can be explained by the fact that these enzymes are also involved in precorrin biosynthesis (Zheng et al. 2016), which is important to produce cobalamin, a cofactor required for the Class II ribonucleotide reductase enzyme.
WL-Pathway (Homoacetogenesis)
In addition to the phylogenetic analysis of the enzymes involved in each of the different pathways for methanogenesis, as well as in the biosynthesis of factor F430, coenzyme M and coenzyme B, we have also studied the distribution of the classical WL pathway (homoacetogenesis). Our analyses indicate that only some bacteria (for instance, Firmicutes-Clostridia, Deltaproteobacteria and Spirochaetes) have the enzymatic machinery necessary for this route (see Fig. S40), which agrees with the current understanding of the metabolic abilities of these organisms (Drake et al. 2008; Shin et al. 2016), whereas in Archaea no lineage has the complete set of enzymes required for homoacetogenesis (Figs. S39 and S41).
Discussion
As suggested by Bapteste et al. (2005), and Williams et al. (2017) and shown by our results, the distribution of hydrogenotrophic enzymes on the Archaea domain (Fig. 2) suggests that this pathway is indeed the oldest methane producing route. Although the first five steps of the hydrogenotrophic pathway correspond to the archaeal MBWL pathway (Fig. 1a), the convergence is at the chemical reaction level, but not at the enzymatic level. In other words, the origin of the hydrogenotrophic route is independent from the autotrophic WL pathway, and the present activity of CO2 fixation in methanogenic-euryarchaeota is an analogous pathway. Moreover, the complete hydrogenotrophic route, including the key Mtr and Mcr enzymes, are exclusively present in a well-defined group of Archaea, which is formed solely by methanogenic Euryarchaeota (Fig. 2). It is important to underline that these analogous but not homologous metabolic activities, provide no evidence of autotrophic or methanogenic abilities of the archaeal ancestor, nor much less of LCA, albeit some metabolic building blocks related to specific cofactor biosynthesis (e.g. tetrahydromethanopterin) could be present in LCA (Chistoserdova 2016; Chistoserdova and Kalyuzhnaya 2018). Our results also are compatible with an independent origin of the enzymatic repertoire for the WL pathway in Bacteria and the hydrogenotrophic methanogenesis in Euryarchaeota as has been suggested by several authors (Martin and Russell 2007; Nitschke and Russell 2013; and references therein).
Maximum likelihood SSU-rRNA phylogeny based on the last tree of life dataset (Hug et al. 2016), in which is shown the distribution of the archaeal methyl branch of Wood-Ljungdahl pathway (MBWL) or otherwise parts thereof (colored squares), as well as the distribution of Bacterial MBWL (stars) and the Mtr and Mcr enzymes (triangles), specifically which homoacetogenesis. Red labels belong to Euryarchaeota phylum. Grey labels represent putative positions of archaeal phyla that was not included in Hug’s analysis. Eukarial branch is colored in orange, archaeal branches in blue and bacterial branches in black. Green circles in middle of the branches are bootstrap values >65%. In the phylogeny TACK makes reference to phyla Thaumarchaeota, Aigarchaeota, Crenarchaeota and Korarchaeota. Fmd Formylmethanofuran dehydrogenase. Ftr Formylmethanofuran-H4MPT formyltransferase. Mch Methenyl-H4MPT cyclohydrolase. Mtd-Hmd H2-forming methylene-H4MPT dehydrogenase. Mer F420-dependent methylene-H4MPT reductase. Mtr Methyltransferase. Mcr Methyl-Coenzyme M Reductase. The letters in the middle of the white circles refer to the different types of methanogenesis, H for Hydrogenotrophic, M for Methylotrophic and A for Acetoclastic
Together with the analysis of the evolution of enzymes with more than one subunit present here, the results suggest a patchwork evolution (Jensen 1976; Ycas 1974) of methanogenesis. Moreover, the phylogenetic distributions of domains and proteins are congruent with the idea that hydrogenotrophic methanogenesis, which can be divided into two classes, is older than acetoclastic and methylotrophic methanogenesis (Bapteste et al. 2005, Fig. 2).
The results presented here also confirm the results of Fournier and Gogarten (2008) on the distribution of Ack (Fig. S23) and Pta (Fig. S24), of the acetoclastic production of methane, and support the suggestion that both genes were transferred from Clostridia to Methanosarcina. We have also shown that Pta and Ack are present in protists, fungi and plants. Neither Ack nor Pta were found in Methanosaeta, confirming other reports (Jetten et al. 1989). Methanosaeta species have an AMP-forming ACS and ADP-forming ACS, that functionally can substitute the Ack and Pta activities (Berger et al. 2012). Although Methanobacterium is reported as a methane producer through hydrogenotrophic methanogenesis, Pta was found in its genetic repertoire, indicating its generalist character in acetate-assimilating metabolisms (see Fig. 1 and Table S1).
The analysis presented here shows that the subunit CdhB is an archaeal innovation (Fig. S26). Each subunit of the acetoclastic CODH/ACS enzyme has a different evolutionary history in Bacteria and Archaea domains, where the distribution of the subunit CdhA is particularly important, because of its involvement in the oxidation activity of the carbonyl group. CdhB is present almost exclusively in Archaea, except for one member of Firmicutes-Clostridia (“Ca. Desulforudis audaxviator”) which apparently has undergone an HGT from Archaea members (Chivian et al. 2008; Bonch-Osmolovskaya 2010). Although, Class I and II CODH/ACS could function in both anabolic acetyl-CoA synthesis and catabolic acetyl-CoA cleavage, their high divergences shown in sequences from CdhC, CdhD subunits (Figs. S42 and S43), and the distinctive rate activity found in different grown conditions (Matschiavelli et al. 2012), suggest a specialization process.
The methylotrophic methanogenesis can be seen like a salvage pathway in which different methylated compounds are integrated to energy metabolism. The simplicity of the route (which requires only methyltransferases and the Mcr enzyme), suggests that this type of methanogenesis arose early in Archaea. However, the distribution of this type of methanogenesis (see Fig. 2 and Table S1) does not support this idea. Moreover, the phylogenetic distributions of MtaA, MtbA, MtsA and MtpA, which participate in the second step on the different methylotrophic pathways, are clearly related to their bacterial counterparts, and can be explained by a series of HGT events (see Figs. S30 and S35–37).
The high frequency of HGT found in many of the subunits and enzymes present in methanogenic pathways from non-methanogenic archaea, bacteria and in some cases even eukarya, suggest that these proteins can participate in other metabolic processes. This appears to be the case of the tungsten-containing formylmethanofuran dehydrogenase subunit E (FwdE) which, in addition of methanogens, is conserved in acetogens where its function is unclear but may have a DNA-binding property as indicated by Shin et al. 2016.
Concluding Remarks
As shown here and in other works (Bapteste et al. 2005; Liu and Whitman 2008; Williams et al. 2017), the distribution and phylogeny of the enzymes that catalyze methane production clearly suggests that the hydrogenotrophic pathway is older than the acetoclastic and methylotrophic routes. Although the evolution of methanogenesis is not free from HGT, the key enzymes Mtr and Mcr and the hydrogenotrophic route are complete only in Euryarchaeota members. However, in the case of the Mcr enzyme they are also present in “Ca. Bathyarchaeota” and “Ca. Verstraetearchaeota”. Furthermore, the distribution of the biosynthetic proteins of the essential coenzymes of the methanogenesis process (for example factor F430, coenzyme M and coenzyme B), is also very peculiar, as they are found only in the Euryarchaeota phylum (see Fig. 3). In consequence, as shown in this work, the distribution of enzymes contradicts proposals of a methanogenic last archaeal common ancestor (Borrel et al. 2016) and do not support the idea that this pathway evolved during the very early stages of life. In other words, methanogenesis is clearly an ancient metabolism but not a truly primordial one. Moreover, the outcome of the analysis of distribution of the methanogenic coenzymes and their biosynthetic enzymes question the methanogenic abilities of some members belonging to “Ca. Bathyarchaeota” and also some belonging to “Ca. Verstraetearchaeota” (see Fig. S44).
References
Adam PS, Borrel G, Gribaldo S (2018) Evolutionary history of carbon monoxide dehydrogenase/acetyl-CoA synthase, one of the oldest enzymatic complexes. Proc Natl Acad Sci U S A 201716667:E1166–E1173. https://doi.org/10.1073/pnas.1716667115
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402. https://doi.org/10.1093/nar/25.17.3389
Bapteste É, Brochier C, Boucher Y (2005) Higher-level classification of the archaea: evolution of methanogenesis and methanogens. Archaea 1(2002):353–363. https://doi.org/10.1155/2005/859728
Becerra A, Delaye L, Islas S, Lazcano A (2007) The very early stages of biological evolution and the nature of the last common ancestor of the three major cell domains. Annu Rev Ecol Evol Syst 38(1):361–379. https://doi.org/10.1146/annurev.ecolsys.38.091206.095825
Becerra A, Rivas M, García-Ferris C, Lazcano A, Peretó J (2014) A phylogenetic approach to the early evolution of autotrophy: the case of the reverse TCA and the reductive acetyl-CoA pathways. Int Microbiol 17:91–97. https://doi.org/10.2436/20.1501.01.211
Berg I, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hügler M, Alber B, Fuchs G (2010) Autotrophic carbon fixation in archaea. Nat Rev Microbiol 8(6):447–460. https://doi.org/10.1038/nrmicro2365
Berger S, Welte C, Deppenmeier U (2012) Acetate activation in Methanosaeta thermophila: characterization of the key enzymes pyrophosphatase and acetyl-CoA Synthetase. Archaea 2012:1–10. https://doi.org/10.1155/2012/315153
Bonch-Osmolovskaya EA (2010) High-temperature deep-subsurface microbial communities as a possible equivalent of ancient ecosystems. Paleontol J 44(7):851–859. https://doi.org/10.1134/s0031030110070130
Borrel G, O’Toole PW, Harris HMB, Peyret P, Brugère J-F, Gribaldo S (2013) Phylogenomic data support a seventh order of methylotrophic methanogens and provide insights into the evolution of Methanogenesis. Genome Biol Evol 5(10):1769–1780. https://doi.org/10.1093/gbe/evt128
Borrel G, Adam PS, Gribaldo S (2016) Methanogenesis and the wood-Ljungdahl pathway: an ancient, versatile, and fragile association. Genome Biol Evol 8(6):1706–1711. https://doi.org/10.1093/gbe/evw114
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. https://doi.org/10.1093/bioinformatics/btp348
Chistoserdova L (2016) Wide distribution of genes for tetrahydromethanopterin/methanofuran-linked C1 transfer reactions argues for their presence in the common ancestor of bacteria and archaea. Front Microbiol 7(SEP):1–5. https://doi.org/10.3389/fmicb.2016.01425
Chistoserdova L, Kalyuzhnaya MG (2018) Current Trends in Methylotrophy. Trends Microbiol xx:1–12. https://doi.org/10.1016/j.tim.2018.01.011
Chivian D, Brodie EL, Alm EJ, Culley DE, Dehal PS, DeSantis TZ, Gihring TM, Lapidus A, Lin L-H, Lowry SR, Moser DP, Richardson PM, Southam G, Wanger G, Pratt LM, Andersen GL, Hazen TC, Brockman FJ, Arkin AP, Onstott TC (2008) Environmental genomics reveals a single-species ecosystem deep within earth. Science 322(5899):275–278. https://doi.org/10.1126/science.1155495
Costa KC, Leigh JA (2014) Metabolic versatility in methanogens. Curr Opin Biotechnol 29(1):70–75. https://doi.org/10.1016/j.copbio.2014.02.012
Drake HL, Gößner AS, Daniel SL (2008) Old acetogens, new light. Ann N Y Acad Sci 1125:100–128. https://doi.org/10.1196/annals.1419.016
Drevland RM, Jia Y, Palmer DRJ, Graham DE (2008) Methanogen homoaconitase catalyzes both hydrolyase reactions in coenzyme B biosynthesis. J Biol Chem 283(43):28888–28896. https://doi.org/10.1074/jbc.M802159200
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797. https://doi.org/10.1093/nar/gkh340
Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD, Tyson GW (2015) Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350(6259):434–438. https://doi.org/10.1126/science.aac7745
Ferry JG (1992) Methane from acetate. J Bacteriol 174(17):5489–5495. https://doi.org/10.1128/jb.174.17.5489-5495.1992
Ferry JG (1993) Methanogenesis: ecology, physiology, biochemistry and genetics. Springer US, Boston. https://doi.org/10.1007/978-1-4615-2391-8
Ferry JG (1997) Enzymology of the fermentation of acetate to methane by Methanosarcina thermophila. Biofactors (Oxford, England) 6(1):25–35. https://doi.org/10.1002/biof.5520060104
Ferry JG (1999) Enzymology of one-carbon metabolism in methanogenic pathways. FEMS Microbiol Rev 23(1):13–38. https://doi.org/10.1111/j.1574-6976.1999.tb00390.x
Ferry JG (2010) How to make a living by exhaling methane. Annu Rev Microbiol 64:453–473. https://doi.org/10.1146/annurev.micro.112408.134051
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39(Web Server issue):W29–W37. https://doi.org/10.1093/nar/gkr367
Fischer JD, Holliday GL, Rahman SA, Thornton JM (2010) The structures and physicochemical properties of organic cofactors in biocatalysis. J Mol Biol 403(5):803–824. https://doi.org/10.1016/j.jmb.2010.09.018
Fournier GP, Gogarten JP (2008) Evolution of acetoclastic methanogenesis in Methanosarcina via horizontal gene transfer from cellulolytic clostridia. J Bacteriol 190(3):1124–1127. https://doi.org/10.1128/JB.01382-07
Fuchs G (2011) Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu Rev Microbiol 65(1):631–658. https://doi.org/10.1146/annurev-micro-090110-102801
Gao B, Gupta RS (2007) Phylogenomic analysis of proteins that are distinctive of archaea and its main subgroups and the origin of methanogenesis. BMC Genomics 8:86. https://doi.org/10.1186/1471-2164-8-86
Graham DE, White RH (2002) Elucidation of methanogenic coenzyme biosyntheses: from spectroscopy to genomics. Nat Prod Rep 19(2):133–147. https://doi.org/10.1039/B103714P
Gribaldo S, Brochier-Armanet C (2006) The origin and evolution of archaea: a state of the art. Philos Trans R Soc Lond Ser B Biol Sci 361(1470):1007–1022. https://doi.org/10.1098/rstb.2006.1841
He Y, Li M, Perumal V, Feng X, Fang J, Xie J, Sievert SM, Wang F (2016) Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat Microbiol 1(6):16035. https://doi.org/10.1038/nmicrobiol.2016.35
Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF (2016) A new view of the tree of life. Nat Microbiol 1(5):16048. https://doi.org/10.1038/nmicrobiol.2016.48
Hügler M, Sievert SM (2011) Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Annu Rev Mar Sci 3:261–289. https://doi.org/10.1146/annurev-marine-120709-142712
Jensen RA (1976) Enzyme recruitment in evolution of new function. Annu Rev Microbiol 30(1):409–425. https://doi.org/10.1146/annurev.mi.30.100176.002205
Jetten MS, Stams AJ, Zehnder AJ (1989) Isolation and characterization of acetyl-coenzyme a synthetase from Methanothrix soehngenii. J Bacteriol 171(10):5430–5435. https://doi.org/10.1128/jb.171.10.5430-5435.1989
Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M (2014) Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res 42(D1):199–205. https://doi.org/10.1093/nar/gkt1076
Kobashi N, Nishiyama M, Tanokura M (1999) Aspartate kinase-independent lysine synthesis in an extremely thermophilic bacterium, Thermus thermophilus: lysine is synthesized via alpha-aminoadipic acid not via diaminopimelic acid. J Bacteriol 181(6):1713–1718
Laso-Pérez R, Wegener G, Knittel K, Widdel F, Harding KJ, Krukenberg V, Meier DV, Richter M, Tegetmeyer HE, Riedel D, Richnow H-H, Adrian L, Reemtsma T, Lechtenfeld OJ, Musat F (2016) Thermophilic archaea activate butane via alkyl-coenzyme M formation. Nature 539(7629):396–401. https://doi.org/10.1038/nature20152
Lessner DJ (2009) Methanogenesis biochemistry. In: eLS. John Wiley & Sons Ltd, Chichester. 1:1–11. https://doi.org/10.1002/9780470015902.a0000573.pub2
Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44(W1):W242–W245. https://doi.org/10.1093/nar/gkw290
Lindahl PA, Chang B (2001) The evolution of acetyl-CoA synthase. Orig Life Evol Biosph 31(4–5):403–434. https://doi.org/10.1023/A:1011809430237
Liu Y, Whitman WB (2008) Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann N Y Acad Sci 1125(1):171–189. https://doi.org/10.1196/annals.1419.019
Mall A, Sobotta J, Huber C, Tschirner C, Kowarschik S, Bačnik K, Mergelsberg M, Boll M, Hügler M, Eisenreich W, Berg IA (2018). Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Science Feb 2;359(6375):563–567. doi: https://doi.org/10.1126/science.aao2410
Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y et al (2012) IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. https://doi.org/10.1093/nar/gkr1044
Martin W, Russell MJ (2007) On the origin of biochemistry at an alkaline hydrothermal vent. Philos Trans R Soc Lond Ser B Biol Sci 362(1486):1887–1925. https://doi.org/10.1098/rstb.2006.1881
Matschiavelli N, Oelgeschläger E, Cocchiararo B, Finke J, Rother M (2012) Function and regulation of isoforms of carbon monoxide dehydrogenase/acetyl coenzyme a synthase in Methanosarcina acetivorans. J Bacteriol 194(19):5377–5387. https://doi.org/10.1128/JB.00881-12
Minh BQ, Nguyen MAT, Von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30(5):1188–1195. https://doi.org/10.1093/molbev/mst024
Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32(1):268–274. https://doi.org/10.1093/molbev/msu300
Nitschke W, Russell MJ (2013) Beating the acetyl coenzyme A-pathway to the origin of life. Phil Trans R Soc B 368:20120258. https://doi.org/10.1098/rstb.2012.0258
Nunoura T, Chikaraishi Y, Izaki R, Suwa T, Sato T, Harada T, Mori K, Kato Y, Miyazaki M, Shimamura S, Yanagawa K, Shuto A, Ohkouchi N, Fujita N, Takaki Y, Atomi H, Takai KA (2018). Primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science Feb 2;359(6375):559–563. doi:https://doi.org/10.1126/science.aao3407
Peretó J (2012) Out of fuzzy chemistry: from prebiotic chemistry to metabolic networks. Chem Soc Rev 41(16):5394–5403. https://doi.org/10.1039/C2CS35054H
Peretó JG, Velasco AM, Becerra A, Lazcano A (1999) Comparative biochemistry of CO2 fixation and the evolution of autotrophy. Int Microbiol 2(1):3–10
Pruitt KD, Tatusova T, Maglott DR (2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35(SUPPL. 1):61–65. https://doi.org/10.1093/nar/gkl842
Shin J, Song Y, Jeong Y, Cho BK (2016) Analysis of the Core genome and pan-genome of autotrophic Acetogenic Bacteria. Front Microbiol 7(Sep 28):1531. https://doi.org/10.3389/fmicb.2016.01531
Smith KS, Ingram-Smith C (2007) Methanosaeta, the forgotten methanogen? Trends Microbiol 15(4):150–155. https://doi.org/10.1016/j.tim.2007.02.002
Sorokin DY, Makarova KS, Abbas B, Ferrer M, Golyshin PN, Galinski EA, Ciordia S, Mena MC, Merkel AY, Wolf YI, van Loosdrecht MCM, Koonin EV (2017) Discovery of extremely halophilic, methyl-reducing euryarchaea provides insights into the evolutionary origin of methanogenesis. Nat Microbiol 2:17081. https://doi.org/10.1038/nmicrobiol.2017.81
Sousa FL, Martin WF (2014) Biochemical fossils of the ancient transition from geoenergetics to bioenergetics in prokaryotic one carbon compound metabolism. BBA Bioenergetics 1837(7):964–981. https://doi.org/10.1016/j.bbabio.2014.02.001
Sousa FL, Thiergart T, Landan G, Nelson-Sathi S, Pereira I, Allen JF, Lane N, Martin WF (2013) Early bioenergetic evolution. Philos Trans R Soc Lond Ser B Biol Sci 368(1622):20130088. https://doi.org/10.1098/rstb.2013.0088
Spang A, Caceres EF, Ettema TJG (2017) Genomic exploration of the diversity, ecology, and evolution of the archaeal domain of life. Science 357(6351):eaaf3883. https://doi.org/10.1126/science.aaf3883
Timmers PHA, Welte CU, Koehorst JJ, Plugge CM, Jetten MSM, Stams AJM (2017) Reverse Methanogenesis and respiration in Methanotrophic archaea. Archaea 2017:1–22. https://doi.org/10.1155/2017/1654237
Ueno Y, Yamada K, Yoshida N, Maruyama S, Isozaki Y (2006) Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature 440(7083):516–519. https://doi.org/10.1038/nature04584
Vanwonterghem I, Evans PN, Parks DH, Jensen PD, Woodcroft BJ, Hugenholtz P and Tyson GW (2016). Methylotrophic methanogenesis discovered in the novel archaeal phylum Verstraetearchaeota. Nature (October):1–9. https://doi.org/10.1038/nmicrobiol.2016.170
Weiss MC, Sousa FL, Mrnjavac N, Neukirchen S, Roettger M, Nelson-Sathi S, Martin WF (2016) The physiology and habitat of the last universal common ancestor. Nat Microbiol 1(9):1–8. https://doi.org/10.1038/nmicrobiol.2016.116
Whitman WB, Bowen TL and Boone DR (2006). The methanogenic Bacteria. In M Dworkin, S Falkow, E Rosenberg, K-H Schleifer and E Stackebrandt (Eds.) The prokaryotes volume 3: archaea. Bacteria: Firmicutes, Actinomycetes (pp. 165–207). Springer New York. https://doi.org/10.1007/0-387-30743-5_9
Williams TA, Szöllősi GJ, Spang A, Foster PG, Heaps SE, Boussau B, Ettema TJG, Embley TM (2017) Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proc Natl Acad Sci U S A 114(23):E4602–E4611. https://doi.org/10.1073/pnas.1618463114
Wolfe JM, Fournier GP (2018) Horizontal gene transfer constrains the timing of methanogen evolution. Nat Ecol Evol 2(5):897–903. https://doi.org/10.1038/s41559-018-0513-7
Woodcroft BJ, Boyd JA, Tyson GW (2016) OrfM: a fast open reading frame predictor for metagenomic data. Bioinformatics 32(17):2702–2703. https://doi.org/10.1093/bioinformatics/btw241
Ycas M (1974) On earlier states of the biochemical system. J Theor Biol 44(1):145–160. https://doi.org/10.1016/S0022-5193(74)80035-4
Zheng K, Ngo PD, Owens VL, Yang X, Mansoorabadi SO (2016) The biosynthetic pathway of coenzyme F430 in methanogenic and methanotrophic archaea. Science 354(6310):339–342. https://doi.org/10.1126/science.aag2947
Zheng Y, Harris DF, Yu Z, Fu Y, Poudel S, Ledbetter RN, Fixen KR, Yang ZY, Boyd ES, Lidstrom ME, Seefeldt LC, Harwood CS (2018). A pathway for biological methane production using bacterial iron-only nitrogenase. Nat Microbiol 3(3):281–286. doi: https://doi.org/10.1038/s41564-017-0091-5
Acknowledgments
We are indebted to Dr. José Alberto Campillo-Balderas for his help with the manuscript. Financial support of DGAPA-UNAM (PAPIIT-IN223916) is gratefully acknowledged. IM-V is a student from the Posgrado en Ciencias Biológicas, Universidad Nacional Autónoma de México (UNAM) and received fellowship 415961 from CONACyT. CGF and JP acknowledge financial support from Mineco/FEDER (grant references: BFU2015-64322-C2-1-R and BIO2015-66960-C3-1-R) and the Generalitat Valenciana (grant reference: PROMETEOII/2014/065).
Author information
Authors and Affiliations
Contributions
Authors contributed equally to this manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Muñoz-Velasco, I., García-Ferris, C., Hernandez-Morales, R. et al. Methanogenesis on Early Stages of Life: Ancient but Not Primordial. Orig Life Evol Biosph 48, 407–420 (2018). https://doi.org/10.1007/s11084-018-9570-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11084-018-9570-9