Abstract
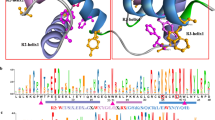
The MYB–CC gene family encode proteins that harbor a combination of characteristic myeloblastosis (MYB) and coiled–coil (CC) domain structures. Some MYB-CC genes have been demonstrated to represent transcription factors regulating phosphate uptake and controlling the starvation response in plants. Despite their physiological importance, a systematic analysis of MYB-CC genes has not been reported in maize. In our study, we identified and characterized maize MYB-CC genes at whole-genome level. A total of 12 maize MYB-CC genes (ZmMYB-CC1 to ZmMYB-CC12) were identified located in six out of the 10 chromosomes of maize. Their gene structures showed similar splicing patterns and large variations of intron length. Multiple sequence alignments revealed that all MYB-CC proteins in maize shared conserved sequence cores corresponding to the MYB and CC domains, respectively. The family expanded in maize partly due to tandem and segmental duplication events. Phylogenetic analysis of MYB-CC genes indicated that the MYB-CC gene family can be divided into two subfamilies and that gene members with same functions were found in the same groups. Results provide a very useful reference for cloning and functional analysis of PHR-like genes in maize and suggest a method to predict and select appropriate candidate genes for functional genomic analysis of useful traits in crop plants.





Similar content being viewed by others
References
Bari R, Pant BD, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141:988–999
Coelho GTCP, Carneiro NP, Karthikeyan AS, Raghothama KG, Schaffert RE, Brandão RL, Paiva LV, Souza IRP, Alves VM, Imolesi A, Carvalho CHS, Carneiro AA (2010) A phosphate transporter promoter from Arabidopsis thaliana AtPHT1;4 gene drives preferential gene expression in transgenic maize roots under phosphorus starvation. Plant Mol Biol Rep 28:717–723
Dai X, You C, Wang L, Chen G, Zhang Q, Wu C (2009) Molecular characterization, expression pattern, and function analysis of the OsBC1L family in rice. Plant Mol Biol 71:469–481
Ding X, Hou X, Xie K, Xiong L (2009) Genome-wide identification of BURP domain-containing genes in rice reveals a gene family with diverse structures and responses to abiotic stresses. Planta 230:149–163
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39:1033–1037
Gollan PJ, Bhave M (2010) Genome-wide analysis of genes encoding FK506-binding proteins in rice. Plant Mol Biol 72:1–16
Griffith AJF, Gelbart WM, Miller JH, Lewontin RC (1999) Chromosomal rearrangements. In: Modern genetic analysis. W.H. Freeman Publishers, New York
Guo AY, Zhu QH, Chen X, Luo JC (2007) GSDS: a gene structure display server. Yi Chuan 29:1023–1026
Huang J, Zhao X, Yu H, Ouyang Y, Wang L, Zhang Q (2009) The ankyrin repeat gene family in rice: genome-wide identification, classification and expression profiling. Plant Mol Biol 71:207–226
Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310–2314
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lipsick JS (1996) One billion years of Myb. Oncogene 13:223–235
Ma K, Xiao J, Li X, Zhang Q, Lian X (2009) Sequence and expression analysis of the C3HC4-type RING finger gene family in rice. Gene 444:33–45
Moore RC, Purugganan MD (2003) The early stages of duplicate gene evolution. Proc Natl Acad Sci USA 100:15682–15687
Nilsson L, Müller R, Nielsen TH (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30:1499–1512
Ren F, Guo Q-Q, Chang L-L, Chen L, Zhao C-Z, Zhong H, Li X-B (2012) Brassica napus PHR1 gene encoding a MYB-like protein functions in response to phosphate starvation. PLoS One 7:e44005
Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15:2122–2133
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schnable PS, Ware D, Fulton RS et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Wang Y, Deng D, Bian Y, Lv Y, Xie Q (2010) Genome-wide analysis of primary auxin-responsive Aux/IAA gene family in maize (Zea mays L.). Mol Biol Rep 37:3991–4001
Wang X, Bai J, Liu H, Sun Y, Shi X, Ren Z (2013) Overexpression of a maize transcription factor ZmPHR1 improves shoot inorganic phosphate content and growth of Arabidopsis under low-phosphate conditions. Plant Mol Biol Rep 31:665–677
Xu P, Zhu HL, Xu HB et al (2010) Composition and phylogenetic analysis of wheat cryptochrome gene family. Mol Biol Rep 37:825–832
Yang L, Zhu H, Guo W, Zhang T (2010) Molecular cloning and characterization of five genes encoding pentatricopeptide repeat proteins from Upland cotton (Gossypium hirsutum L.). Mol Biol Rep 37:801–808
Yue G, Hu X, He Y, Yang A, Zhang J (2010) Identification and characterization of two members of the FtsH gene family in maize (Zea mays L.). Mol Biol Rep 37:855–863
Zhou J, Jiao FC, Wu ZC, Li YY, Wang XM, He XW, Zhong WQ, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146:1673–1686
Acknowledgements
The Major Transgenic Organism Breeding Project initiative was supported by grants from the Chinese Ministry of Agriculture (2009ZX08003 and 2011ZX08003) and Shanxi International Cooperation Project (2012081005) to JB. FJS was supported by the Educational and Professional Leave from Georgia Gwinnett College when he participated in this work. Research at Illinois was supported by USDA National Institute of Food and Agriculture, Hatch Project 1014249 and a Blue Waters supercomputer allocation to GCA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Bai, J., Sun, F., Wang, M. et al. Genome-wide analysis of the MYB-CC gene family of maize. Genetica 147, 1–9 (2019). https://doi.org/10.1007/s10709-018-0042-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-018-0042-y