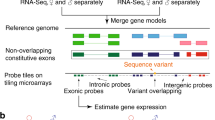
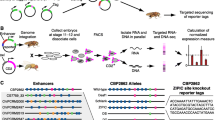
Abstract
Gene regulatory networks control the complex programs that drive development. Deciphering the connections between transcription factors (TFs) and target genes is challenging, in part because TFs bind to thousands of places in the genome but control expression through a subset of these binding events. We hypothesize that we can combine natural variation of expression levels and predictions of TF binding sites to identify TF targets. We gather RNA-seq data from 71 genetically distinct F1 Drosophila melanogaster embryos and calculate the correlations between TF and potential target genes' expression levels, which we call “regulatory strength.” To separate direct and indirect TF targets, we hypothesize that direct TF targets will have a preponderance of binding sites in their upstream regions. Using 14 TFs active during embryogenesis, we find that 12 TFs showed a significant correlation between their binding strength and regulatory strength on downstream targets, and 10 TFs showed a significant correlation between the number of binding sites and the regulatory effect on target genes. The general roles, e.g. bicoid’s role as an activator, and the particular interactions we observed between our TFs, e.g. twist’s role as a repressor of sloppy paired and odd paired, generally coincide with the literature.
Similar content being viewed by others
References
E. H. Davidson, Nature 468, 911 (2010).
I. S. Peter and E. H. Davidson, Cell 144, 970 (2011).
X. Li, S. MacArthur, R. Bourgon, et al., PLoS Biol. 6, e27 (2008).
J. M. Vaquerizas, S. K. Kummerfeld, S. A. Teichmann, and N. M. Luscombe, Nat. Rev. Genet. 10, 252 (2009).
S. V. Nuzhdin, A. Rychkova, and M. W. Hahn, Trends Genet. 26, 51 (2010).
S. Yang, H. K. Yalamanchili, X. Li, et al., Bioinformatics 27 (21), 2972 (2011).
M. Levo and E. Segal, Nat. Rev. Genet. 15, 453 (2014).
A. Nasiadka, B. H. Dietrich, and H. M. Krause, Adv. Dev. Biol. Biochem. 12, 155 (2002).
S. Bonn and E. E. Furlong, Curr. Opin. Genet. Dev. 18, 513 (2008).
M. Levine and E. H. Davidson, Proc. Natl. Acad. Sci. U. S. A. 102, 4936 (2005).
K. Howard and P. Ingham, Cell 44, 949 (1986).
C. M. Bergman, J. W. Carlson, and S. E. Celniker, Bioinformatics 21, 1747 (2005).
R. Bonneau, Nat. Chem. Biol. 4, 658 (2008).
B. W. Busser, M. L. Bulyk, and A. M. Michelson, Curr. Opin. Genet. Dev. 18, 521 (2008).
R. E. Bumgarner and K. Y. Yeung, Comput. Syst. Biol. 541, 225 (2009).
P. J. Park, Nat. Rev. Genet. 10, 669 (2009).
S. Pepke, B. Wold, and A. Mortazavi, Nat. Methods 6, S22 (2009).
Y. R. Wang and H. Huang, J. Theor. Biol. 362, 53 (2014).
D. J. Kliebenstein, Plant Syst. Biol. 553, 227 (2009).
J. A. Lewis, I. M. Elkon, M. A. McGee, et al., Genetics 186, 1197 (2010).
S. Mostafavi, A. Ortiz-Lopez, M. A. Bogue, et al., J. Immunol. 193, 4485 (2014).
S. V. Nuzhdin, D. M. Tufts, and M. W. Hahn, Evol. Dev. 10, 683 (2008).
W. Jin, R. M. Riley, R. D. Wolfinger, et al., Nat. Genet. 29, 389 (2001).
S. V. Nuzhdin, M. L. Wayne, K. L. Harmon, and L. M. McIntyre, Mol. Biol. Evol. 21, 1308 (2004).
S. V. Nuzhdin, M. L. Friesen, and L. M. McIntyre, Trends Genet. 28, 421 (2012).
T. F. Mackay, S. Richards, E. A. Stone, et al., Nature 482, 173 (2012).
D. Campo, K. Lehmann, C. Fjeldsted, et al., Mol. Ecol. 22, 5084 (2013).
J. P. Dunham and M. L. Friesen, Cold Spring Harb. Protoc. 9, 820 (2013).
A. Dobin, C. A. Davis, F. Schlesinger, et al., Bioinformatics 29, 15 (2013).
S. Anders, P. T. Pyl, and W. Huber, Bioinformatics 31 (2), 166 (2015).
A. Mortazavi, B. A. Williams, K. McCue, et al., Nat. Methods 5, 621 (2008).
T. Sandmann, C. Girardot, M. Brehme, et al., Genes Dev. 21, 436 (2007).
J. A. Campos-Ortega and V. Hartenstein, The Embryonic Development of Drosophila melanogaster (Springer Sci. Business Media, 2013).
S. Thomas, X.-Y. Li, P. J. Sabo, et al., Genome Biol. 12, R43 (2011).
G. Z. Hertz and G. D. Stormo, Bioinformatics 15, 563 (1999).
L. J. Zhu, R. G. Christensen, M. Kazemian, et al., Nucleic Acids Res. 39, D111 (2011).
Y. Benjamini and Y. Hochberg, J. R. Stat. Soc. Series B 57 (1), 289 (1995).
Z. Wang, M. Gerstein, and M. Snyder, Nat. Rev. Genet. 10, 57 (2009).
G. Struhl, K. Struhl, and P. M. Macdonald, Cell 57, 1259 (1989).
M. Leptin, Genes Dev. 5, 1568 (1991).
R. M. Cripps, B. L. Black, B. Zhao, et al., Genes Dev. 12, 422 (1998).
A. Stathopoulos, M. Van Drenth, A. Erives, et al., Cell 111, 687 (2002).
M. D. Schroeder, M. Pierce, J. Fak, et al., PLoS Biol. 2, e271 (2004).
J. Zeitlinger, R. P. Zinzen, A. Stark, et al., Genes Dev. 21, 385 (2007).
A. Ochoa-Espinosa, D. Yu, A. Tsirigos, et al., Proc. Natl. Acad. Sci. U. S. A. 106, 3823 (2009).
A. Porcher and N. Dostatni, Curr. Biol. 20, R249 (2010).
X. Wu, R. Vakani, and S. Small, Development 125, 3765 (1998).
G. F. Hewitt, et al., Development 126, 1201 (1999).
E. Morán and G. Jiménez, Mol. Cell. Biol. 26, 3446 (2006).
J. O. Yáñez-Cuna, E. Z. Kvon, and A. Stark, Trends Genet. 29, 11 (2013).
A. S. Manoukian and H. M. Krause, Genes Dev. 6, 1740 (1992).
S. Barolo and M. Levine, EMBO J. 16, 2883 (1997).
G. Jiménez, Z. E. Paroush, and D. Ish-Horowicz, Genes Dev. 11, 3072 (1997).
M. Kobayashi, R. E. Goldstein, M. Fujioka et al., Development 128, 1805 (2001).
M. Fujioka, G. L. Yusibova, N. H. Patel, et al., Development 129, 4411 (2002).
D. Bianchi-Frias, A. Orian, J. J. Delrow, et al., PLoS Biol. 2, e178. (2004).
Y. Hiromi and W. J. Gehring, Cell 50, 963 (1987).
S. G. Kramer, T. M. Jinks, P. Schedl, and J. P. Gergen, Development 126, 191 (1999).
Y. Yu, M. Yussa, J. Song, et al., Mech. Dev. 83, 95 (1999).
A. Nasiadka, A. Grill, and H. M. Krause, Development 127, 2965 (2000).
J. C. Wheeler, K. Shigesada, J. P. Gergen, and Y. Ito, Semin. Cell Dev. Biol. 11, 369 (2000).
P. I. Zuo, D. Stanojević, J. Colgan, et al., Genes Dev. 5, 254 (1991).
M. V. Staller, B. J. Vincent, M. D. J. Bragdon, et al., Proc. Natl. Acad. Sci. U. S. A. 112, 785 (2015).
F. Sauer and H. Jäckle, Nature 353, 563 (1991).
F. Sauer, J. D. Fondell, Y. Ohkuma, et al., Nature 375, 162 (1995).
A. La Rosée-Borggreve, T. Häder, D. Wainwright, et al., Mech. Dev. 89, 133 (1999).
J. Heemskerk, S. DiNardo, R. Kostriken, and P. H. O’Farrell, Nature 352, 404 (1991).
T. Tabata, S. Eaton, and T. B. Kornberg, Genes Dev. 6, 2635 (1992).
C. Alexandre and J. P. Vincent, Development 130, 729 (2003).
M. Rembold, L. Ciglar, J. O. Yáñez-Cuna, et al., Genes Dev. 28, 167 (2014).
L. Sánchez and D. Thieffry, J. Theor. Biol. 211, 115 (2001).
J. Jaeger, M. Blagov, D. Kosman, et al., Genetics 167 (4), 1721 (2004).
J. Jaeger, Cell. Mol. Life Sci. 68, 243 (2011).
F. Liu, A. H. Morrison, and T. Gregor, Proc. Natl. Acad. Sci. U. S. A. 110, 6724 (2013).
I. A. Gula and A. M. Samsonov, Bioinformatics 31, 714 (2015).
S. MacArthur, X.-Y. Li, J. Li, et al., Genome Biol. 10, R80 (2009).
J. M. Franco-Zorrilla, I. López-Vidriero, J. L. Carrasco, et al., Proc. Natl. Acad. Sci. U. S. A. 111, 2367 (2014).
B. R. Graveley, A. N. Brooks, J. W. Carlson, et al., Nature 471 (7339), 473 (2011).
A. S. Hammonds, C. A. Bristow, W. W. Fisher, et al., Genome Biol. 14, R140 (2013).
M. D. Biggin and R. Tjian, Funct. Integr. Genomics 1, 223 (2001).
M. Levine and R. Tjian, Nature 424, 147 (2003).
S. Björklund, G. Almouzni, I. Davidson, et al., Cell 96, 759 (1999).
A. Tanay, Genome Res. 16, 962 (2006).
M. Mannervik, Y. Nibu, H. Zhang, and M. Levine, Science 284, 606 (1999).
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
Supplementary materials are available for this article at 10.1134/S0006350918010128 and are accessible for authorized users.
Rights and permissions
About this article
Cite this article
Osman, N.M., Kitapci, T.H., Vlaho, S. et al. Inference of Transcription Factor Regulation Patterns Using Gene Expression Covariation in Natural Populations of Drosophila melanogaster. BIOPHYSICS 63, 43–51 (2018). https://doi.org/10.1134/S0006350918010128
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006350918010128