Introduction
Robotic spacecraft at Mars have found soils that are rich in salts, with a high abundance of sulphur compounds present as sulphates of Mg, Ca and some Fe (Clark & van Hart Reference Clark and van Hart1981; Clark Reference Clark1993; Clark et al. Reference Clark, Morris, McLennan, Gellert, Jolliff, Knoll, Squyres, Lowenstein, Ming and Tosca2005; Wänke et al. Reference Wänke, Brückner, Dreibus, Rieder and Ryabchikov2001; Altheide et al. Reference Altheide, Chevrier, Nicholson and Denson2009; McKay et al. Reference McKay, Stoker, Glass, Davé, Davila, Heldmann, Marinova, Fairen, Quinn and Zacny2013). Chlorides and perchlorates of Mg, Na or Ca are present, but are significantly less abundant than sulphates (S : Cl ratio of 4 : 1). Heavy brines could form as liquid water is generated from melting permafrost or by deliquescent salts (Cull et al. Reference Cull, Arvidson, Catalano, Ming, Morris, Mellon and Lemmon2010; Lanza et al. Reference Lanza, Meyer, Okubo, Newson and Wiens2010; McEwen et al. Reference McEwen, Ojha, Dundas, Mattson, Byrne, Wray, Cull, Murchie, Thomas and Gulick2011; Möhlmann & Thomsen Reference Möhlmann and Thomsen2011; McKay et al. Reference McKay, Stoker, Glass, Davé, Davila, Heldmann, Marinova, Fairen, Quinn and Zacny2013). Potential brines may be dominated by MgSO4, as Ca and Fe sulphates are sparingly soluble. To live in ephemeral moist areas on Mars, organisms may need to tolerate and grow at high salinities. Furthermore, the major ions may not be Na and Cl, which dominate hypersaline environments on Earth, but rather Mg and sulphate. Understanding microbial life at high MgSO4 concentrations has relevance to the search for life on Mars and other celestial bodies such as Callisto, Enceladus, Europa and Ganymede, where subsurface oceans appear to be salty (Kargel et al. Reference Kargel, Kaye, Head, Marion, Sassen, Crowley, Ballesteros, Grant and Hogenbloom2000; Marion et al. Reference Marion, Fritsen, Eicken and Payne2003; Hussmann et al. Reference Hussmann, Sohl and Spohn2006; Mottl et al. Reference Mottl, Glazer, Kaiser and Meech2007; Postberg et al. Reference Postberg, Schmidt, Hillier, Kempf and Srama2011).
Natural epsomite environments are rare and little experimental microbiology has been performed at saturating concentrations of MgSO4 (Handy Reference Handy1916; Anderson Reference Anderson1958; Markovitz Reference Markovitz1961; Markovitz & Sylvan Reference Markovitz and Sylvan1962; Boring et al. Reference Boring, Kushner and Gibbons1963; Hammer Reference Hammer1978, Reference Hammer1986; Laiz et al. Reference Laiz, Recio, Hermosin, Saiz-Jimenez, Ciferri, Tiano and Mastromei2000; Mandrioli & Saiz-Jimenez Reference Mandrioli and Saiz-Jimenez2002; Nesbitt Reference Nesbitt, Spencer and Chou2004; Hyde et al. Reference Hyde, Foster, King, Southam and Nushaj2007; Foster et al. Reference Foster, King, Hyde and Southam2010; Crisler et al. Reference Crisler, Newville and Schneegurt2009, Reference Crisler, Newville, Chen, Clark and Schneegurt2012; Lindemann et al. Reference Lindemann, Moran, Dohnalkova, Kim, Kennedy, Stolyar, Maors, Wiley, Konopka and Fredrickson2012). Prohibitively high epsomite concentrations have been advanced as potentially the most extreme of the near-surface conditions at Mars (Tosca et al. Reference Tosca, Knoll and McLennan2008). Our recent study screened a diverse collection of halotolerant bacteria from the Great Salt Plains (GSP) of Oklahoma for epsotolerance, the ability to grow at high MgSO4 concentrations (Crisler et al. Reference Crisler, Newville, Chen, Clark and Schneegurt2012). The vast majority of these broadly halotolerant isolates, including Halomonas and Bacillus, grew in the presence of 2 M MgSO4, even though the isolates were collected from salt flats rich in NaCl, not MgSO4. None of the isolates were epsophilic, requiring high MgSO4 concentrations for growth, but none were halophilic, requiring high NaCl concentrations for growth, either (Schneegurt Reference Schneegurt and Vreeland2012). There was not a direct correspondence between the degree of halotolerance and the degree of epsotolerance in individual isolates. The current report extends these observations to bacterial isolates from Hot Lake, naturally rich in MgSO4, mainly as epsomite.
The potential of terrestrial microbes introduced into the Martian environment to grow and replicate as a function of temperature, salt concentration, and other environmental factors is relevant to the robotic exploration of Mars (NASA, 1980; DeVincenzi et al. Reference DeVincenzi, Stabekis and Barengoltz1996; Cooper et al. Reference Cooper, La Duc, Probst, Vaishampayan, Stam, Benardini, Piceno, Anderson and Venkateswaran2011). Given their low humidity (40±5%), the Class 100K clean rooms used as spacecraft assembly facilities (SAFs) may select for stress-tolerant microbes, particularly salinotolerant bacteria, those that potentially pose the greatest threats to planetary protection. Extensive studies of the microbial populations in SAFs have been performed previously using cultivation (Favero et al. Reference Favero, Puleo, Marshall and Oxborrow1968; Favero Reference Favero1971; Foster & Winans Reference Foster and Winans1975; Puleo et al. Reference Puleo, Fields, Bergstrom, Oxborrow, Stabekis and Koukol1977; Stieglmeier et al. Reference Stieglmeier, Wirth, Kminek and Moissl-Eichinger2009; Probst et al. Reference Probst, Vaishampayan, Osman, Moissl-Eichinger, Anderson and Venkateswaran2010) and molecular strategies (Moissl et al. Reference Moissl, Bruckner and Venkateswaran2008; La Duc et al. Reference La Duc, Osman, Vaishampayan, Piceno, Anderson, Spry and Venkateswaran2009). Of the bacterial strains isolated from SAFs, some exhibited growth at 10% NaCl (Venkateswaran et al. Reference Venkateswaran, Satomi, Chung, Kern, Koukol, Basic and White2001, Reference Venkateswaran, Kempf, Chen, Satomi, Nicholson and Kern2003a, Reference Venkateswaran, Hattori, La Duc and Kernb; La Duc et al. Reference La Duc, Nicholson, Kern and Venkateswaran2003; Link et al. Reference Link, Sawyer, Venkateswaran and Nicholson2003; Kempf et al. Reference Kempf, Chen, Kern and Venkateswaran2005). A wide variety of archaea have been observed in SAFs, some of which fall into groups associated with hypersaline environments (Moissl et al. Reference Moissl, Bruckner and Venkateswaran2008). Thus, the potential exists for contamination of spacecraft with halotolerant microbes in SAFs, however, salts in Mars soils are rich in MgSO4, not NaCl, and there might not be a direct correspondence between tolerances to different salts.
The Canadian Plains is an area rich in athalassohaline lakes that have been used for mining sulphate salts including Glauber's salt and epsomite (Hammer Reference Hammer1978, Reference Hammer1986; Haynes & Hammer Reference Haynes and Hammer1978; Last & Slezak Reference Last and Slezak1988; Last & Ginn Reference Last and Ginn2005). A most unique feature of the lakes in this region is that 53 of 60 lakes were dominated by sulphate anions, exceeding 80 eq.% in more than two thirds of the lakes. The earliest microbiological examination of an epsomite lake was limited to descriptive work at Hot Lake, WA by Anderson (Reference Anderson1958), who reported sulphidic green mats in deeper areas, suggestive of Chlorobium, Oscillatoria and Plectonema. Initial deep sequencing of Hot Lake mat communities has shown a predominance of cyanobacteria (Oscillatoria and Phormidium), Chromatia (Halochromatium and Thiohalocapsa), and purple non-sulphur bacteria (Roseobacter and Rhodovulum), along with other organisms involved in the sulphur cycle, Spirochaetes and Clostridia (Lindemann et al. Reference Lindemann, Moran, Dohnalkova, Kim, Kennedy, Stolyar, Maors, Wiley, Konopka and Fredrickson2012). Basque Lake, BC, a related series of epsomite playas, was the examined using IR spectroscopy to detect certain signatures of life (Hyde et al. Reference Hyde, Foster, King, Southam and Nushaj2007; Foster et al. Reference Foster, King, Hyde and Southam2010). While enrichment cultures for halotolerant microbes were positive, no isolates or microbiological analyses were reported.
The current work characterizes the microbial populations of Hot Lake using cultivation and molecular ecology techniques. A salinotolerant isolate collection was captured and curated through phylogenetic analysis of 16S rRNA gene sequences. Isolates were further characterized by testing for epsotolerance and halotolerance. Random clone libraries of bacterial 16S rRNA gene sequences were analysed phylogenetically. The microbial community at epsomite-rich Hot Lake is compared to that of the halite-rich GSP. This is placed in the context of Mars exploration and planetary protection. Preliminary accounts of this work have been presented previously (Crisler et al. Reference Crisler, Kilmer, Rowe, Cunderla, Madu and Schneegurt2010; Kilmer et al. Reference Kilmer, Eberl, Rowe, Cunderla and Schneegurt2011, Reference Kilmer, Eberl, Crisler, Cunderla, Madu and Schneegurt2012).
Materials and methods
Site description and sample collection
Water and lake margin soils (∼100 g from top 4 cm) were collected from Hot Lake, WA (T. 40 N., R. 27 E., W.M., Section 7, SE¼, Section 18, NE¼, Okanogan County, WA; centered at 48°58′24.70″ N 119°28′33.91″ W; Fig. 1) in October 2009 using sterile tools and containers. Duplicate samples were collected, with one aliquot being frozen in the field and kept frozen during transport on dry ice for molecular work and the other aliquot being held fresh for live culture work. The level of the lake was relatively low at the time of sampling, the surface and pore waters were near saturation, margin soils were crusted with epsomite, and the pH was near 8. The lake is surrounded by mature forest in a mountainous area at an elevation of 583 m. The soils were predominantly a grey or black fine clay and silt, often with a strong sulphidic odour, and thin ice was present on the surface of the lake. Pore and lake water salinity was measured using a salinometer and found to be near saturation.

Fig. 1. Map of Hot Lake showing sampling sites for the current study marked as S1–S7. The altitude of the lake surface (583 m) is shown with topographical relief indicating that Hot Lake is in a depression surrounded by hills.
Enrichment, isolation and characterization
Direct plating, liquid enrichment and dilution plating were used to isolate halotolerant and epsotolerant microbes from Hot Lake waters and soils. The media used were based on SP medium (Caton et al. Reference Caton, Witte, Ngyuen, Buchheim, Buchheim and Schneegurt2004), a nutrient-rich and moderately saline (10%) medium containing per litre: NaCl, 98 g; KCl, 2.0 g; MgSO4·7H2O, 1.0 g; CaCl2·2H2O, 0.36 g; NaHCO3, 0.06 g; NaBr, 0.23 g; FeCl3·6H2O, 1.0 mg; trace minerals, 0.5 ml; Bacto tryptone, 5.0 g; yeast extract, 10.0 g; glucose, 1.0 g; final pH 7.0. This was prepared with either 10% NaCl, 10% MgSO4 or 2 M MgSO4. Although these media were useful for liquid enrichments, agar plates with 2 M MgSO4 would not gel sufficiently, so isolates from liquid enrichments in this medium were transferred to medium with 10% NaCl. Enrichment cultures (100 ml) were maintained as shake-flasks on a rotary shaking platform (2.5 cm stroke dia) at 150 rpm and incubated at 7, 25 or 37 °C before aliquots (100 μl) were plated after 24 or 48 h. For dilution plating, samples (10 ml or 10 g) were diluted 10-fold with appropriate media and serially diluted prior to plating. In some cases, soil or water aliquots (approximately 1 ml or 0.5 g) were spread directly onto the surface of plates. The plates were maintained at the appropriate temperature in a moist box and colonies were collected after several days. Colonies were marked and plates kept for several weeks during which time representatives of new colony types were harvested to capture slower growing organisms.
Colonies arising on plates were selected for isolation based on gross morphological and physiological features, including pigmentation, size, margin or rate of growth. Colonies were transferred to fresh SP agar plates with 10% NaCl or 10% MgSO4 and isolated using the streak-plate method. Each isolate was subjected to at least five successive streak-platings to ensure clonal purity; the length of this process ranged among isolates from 2 to 16 wk. The isolates were curated as 50% glycerol stocks at −80 °C and as agar slants.
Isolates were Gram-stained using a Protocol Gram-staining kit (Fisher Diagnostics) following the manufacturer's instructions. SP medium composition was modified with different concentrations of NaCl or MgSO4 to measure salinity tolerances in liquid shake-tubes. Results are reported as the maximum and minimum conditions for growth. A positive threshold of 0.05 OD units was used and inconclusive tests were repeated.
PCR, cloning and DNA sequencing
Crude DNA extracts from each isolate were prepared using a freeze-thaw technique as described in Caton et al. (2004). Genomic DNA in the supernatant was the target of PCR amplification of nearly complete 16S rRNA gene fragments using bacterial primers (EUBPA: 5′-AGAGTTTGATCCTGGCTCAG-3′ and EUBPH: 5′-AAGGAGGTGATCCAGCCGCA-3′) (Edwards et al. Reference Edwards, Rogall, Blöcker, Emde and Böttger1989). PCRs were performed in a thermal cycler (Eppendorf Mastercycler) as 25 μl reactions containing 0.2 μM of each primer, 1 U of ExTaq DNA polymerase and associated master mix (Takara) and 5 μl of cell extract. DNA was denatured at 95 °C for 2 min, followed by 40 cycles of 95 °C for 1 min, 50 °C for 1 min and 72 °C for 1 min, with a final 5 min extension at 72 °C. PCR amplicons were single-pass sequenced at the University of Kansas Biodiversity Institute using the EUBPA primer.
A metagenomic DNA extract was made directly from Hot Lake soil using the protocol of Bürgmann et al. (Reference Bürgmann, Pesaro, Widmer and Zeyer2001), with some modifications as previously described (Caton & Schneegurt Reference Caton and Schneegurt2012). Briefly, cell breakage in soil samples is performed by bead beating with CTAB detergent followed by organic extraction and precipitation with polyethylene glycol. The metagenomic extract was used as the template for PCR amplification of bacterial 16S rRNA gene sequences. Ten separate PCR amplicon populations were pooled following purification by band excision from a 2% agarose gel after electrophoresis. Clone libraries were generated from the pooled amplicons using a TOPO-TA blue-white cloning system in Escherichia coli (Invitrogen) following the manufacturer's instructions. More than 200 clones were randomly collected and inoculated into 96-well plates, with plasmid isolation and single-pass insert sequencing by a commercial vendor (Agencourt) using the EUBPA primer. Initial sequences were trimmed to remove remaining vector regions, leaving sequences of approximately 800 bp for analysis. All sequences appear in GenBank with accession numbers KC705245 to KC705342 for cultured bacterial isolates and KC705343 to KC705470 for uncultured bacterial clones.
Sequence analyses
Sequences were automatically aligned using Clustal-W (Thompson et al. Reference Thompson, Higgins and Gibson1994) and then manually examined and trimmed in MacClade v4.08 (Sinauer Associates). Contextual 16S rRNA gene sequences were identified in GenBank using BLAST (Altschul et al. Reference Altschul, Gish, Miller, Myers and Lipman1990) or by comparison with relevant literature. PAUP 4.0 b10 (Swofford Reference Swofford1998) generated phylogenetic trees using distance analysis with Jukes–Cantor rules and the neighbour-joining algorithm. Sequences were trimmed to equal lengths, with sequences less than 500 bp removed, and positions with gaps and ambiguous bases ignored, giving 500–600 positions for analysis. Bootstrap analysis was used to assess the relative support for each branch with a total of 100 replicates conducted heuristically using the distance-based neighbour-joining algorithm and the nearest-neighbour-interchange algorithm in PAUP. The trees were rooted using Methanospirillum hungatei as the functional outgroup. Putative chimeras (approximately 15% of the sequences) were identified through iterative analyses using Pintail within MOTHUR (Schloss et al. Reference Schloss, Westcott, Ryabin, Hall, Hartmann, Hollister, Lesniewski, Oakley, Parks and Robinson2009), manually examined, and removed if necessary. Full phylogenetic trees with GenBank accession numbers can be found in supplementary materials (Figs. S1–S3) (available at http://journals.cambridge.org/IJA). Distance files were further analysed using the MOTHUR statistical package to determine Chao1 estimators, Simpson indexes, non-parametric Shannon indexes, rarefaction curves and OTUs at various levels of sequence similarity. Library comparisons were made using BLAST.
Results
Collection and identification of bacterial isolates
All seven soil and water samples provided bacterial isolates, with the soil samples from Sites 2, 4 and 7 each providing approximately 25% of the collection. Nearly 100 aerobic heterotrophic bacterial isolates were acquired from Hot Lake by dilution plating and repetitive streaking. Most isolates were obtained from enrichment cultures at room temperature in SP medium supplemented with 10% NaCl. Approximately a third of the isolates were obtained at 30 or 37 °C in SP medium supplemented with 10% or 2 M MgSO4. The enrichment medium and temperature and the sampling site for each isolate are given in the full phylogenetic trees (Figs. S1 and S2). Representatives of the most common colony types from all seven of the soil and water samples were collected, increasing the degree of duplication in the overall collection. Phylogenetic analysis was performed on 16S rRNA gene sequences to identify each isolate (Figs. 2, 3, S1 and S2).

Fig. 2. Phylogenetic tree for Gram-negative bacteria from Hot Lake based on 16S rRNA gene sequences. Bootstrap values greater than 50% are shown. A full tree with GenBank accession numbers, sample locations, and enrichment conditions can be found in Fig. S1.

Fig. 3. Phylogenetic tree for Gram-positive bacteria from Hot Lake based on 16S rRNA gene sequences. Bootstrap values greater than 50% are shown. A full tree with GenBank accession numbers, sample locations, and enrichment conditions can be found in Fig. S2.
Serial dilution plating on SP medium with 10% salinity was used to isolate the most abundant culturable microbes in Hot Lake samples. Fourteen isolates were obtained from the highest positive dilutions (106–108) of these series. Halomonas appears to be the most abundant culturable genera; isolated (44 total isolates) from all seven soil and water samples, with six isolates collected from the highest positive dilution plates (HL 12, 19, 22, 23, 24, 33 and 75). The other abundant Gram-negative genera observed on dilution plates were Idiomarina (HL78) and Marinobacter (HL77). Low G+C Gram-positive Marinococcus were abundant and were isolated from Sites 2, 4 and 7 (17 total isolates), with three isolates collected from the highest positive dilution plates (HL 9, 25 and 27). The other abundant Gram-positive genera observed on dilution plates were low G+C Planococcus (HL1) and the actinomycetes Nocardiopsis (HL79) and Nesterenkonia (HL76). Bacillus species were represented by nine isolates that were all enriched at 30 or 37 °C, but no Bacillus were isolated from the highest positive dilution plates. In addition to these abundant genera, isolates were obtained from Corynebacterium, Exiguobacterium, Kocuria, Staphylococcus and Virgibacillus. No archaea or distinctly red microbes were isolated from Hot Lake despite enrichments in high-salt media at 37 °C. While not described in the current work, a number of cyanobacteria and fungi were cultured at high salinities and anaerobic bottles were positive for fermenters and sulphate-reducers at 10% NaCl and 2 M MgSO4.
Epsotolerance and halotolerance in bacterial isolates
The Hot Lake isolate collection was screened for growth tolerance to high NaCl and MgSO4 concentrations. Nearly all (82%) of the bacterial isolates from Hot Lake grew at 50% MgSO4 (approximately 2 M). More than half (58%) grew at 60% MgSO4, near saturation. Broad epsotolerance was expected at Hot Lake given its seasonal changes in salinity with snowmelt and summer evaporation. Salinity tolerance ranges of representative isolates from each Hot Lake phylotype are shown in Fig. 4 and the results for each isolate are given in Table S1(available at http://journals.cambridge.org/IJA). It is interesting to note that some isolates, such as HL 77 and 78, exhibited poor or no growth above 10% MgSO4. No epsophilic organisms, requiring high MgSO4 concentrations for growth, have been isolated.

Fig. 4. Epsotolerance of Hot Lake bacterial isolates. Bars indicate the ranges permissible for growth.
The Hot Lake isolates showed remarkable growth tolerance to high NaCl concentrations, although their natural environment is not rich in chlorides or monovalent cations. All isolates grew at 10% NaCl and three-quarters of the isolates grew at 20% salinity (∼3.5 M NaCl). Note that the water activity of 20% NaCl (0.82) is substantially lower than that of saturated MgSO4 (0.90). This is due in part to limited dissociation of MgSO4 in solution. Some of the isolates (16%), predominantly Marinococcus, showed growth at 30% NaCl, near saturation. Salinity tolerance ranges of representative isolates from each Hot Lake phylotype are shown in Fig. 5 and the results for each isolate are given in Table S1. There was not always a direct correspondence between the epsotolerance and halotolerance of individual isolates as exemplified by HL 94 and 99 which grow at 60% MgSO4 but do not grow above 15% NaCl (cf. Figs. 4 and 5).

Fig. 5. Halotolerance of Hot Lake bacterial isolates. Bars indicate the ranges permissible for growth and the optimal salinities for growth are indicated by closed squares.
Culture-independent clone library
A bacterial 16S rRNA gene clone library was prepared from direct DNA extracts of the same soil sample (Site 7) that was the source of many isolates from the cultivation campaign. To reduce biases in the library, the amplicons from ten separate PCRs were combined in the cloning reaction. Approximately 200 random bacterial 16S rRNA gene clones were sequenced. Chimeras were removed (Huber et al. Reference Huber, Faulkner and Hugenholtz2004) and a few sequences were too short for analysis. Overall clustering of major monophyletic groups is shown in Figs. 2, 3, 6 and S1–S3 and followed expected patterns. None of the sequences on the Unclassified tree (Figs. 6 and S3) were represented by cultured isolates; only clones from direct DNA extracts were assigned to these clades. A similar approach was applied using archaeal 16S rRNA gene primers (Caton et al. Reference Caton, Caton, Witte and Schneegurt2009). Although archaeal sequences could be detected by PCR, these appear to be in low abundance and a clone library was not pursued.

Fig. 6. Phylogenetic tree for Chloroflexi and unclassified bacteria from Hot Lake based on 16S rRNA gene sequences. Bootstrap values greater than 50% are shown. A full tree with GenBank accession numbers, sample locations, and enrichment conditions can be found in Fig. S3.
Estimation of diversity and coverage
The results of statistical estimates (Schloss et al. Reference Schloss, Westcott, Ryabin, Hall, Hartmann, Hollister, Lesniewski, Oakley, Parks and Robinson2009) of species richness, evenness, and the efficiency of species collection for the culture-independent clone library are given in Table 1 and Fig. 7. The data are presented at several levels of sequence identity, reflecting commonly used thresholds for the taxonomic levels of strain (99%), species (97%), genus (94%) and division (88%). The number of OTUs increases at higher levels of sequence identity as expected. Using a threshold of 99% sequence identity, 84 OTUs were identified within the bacterial sequences. Good's coverage values were relatively high and consistent at all levels of identity (Schloss et al. Reference Schloss, Westcott, Ryabin, Hall, Hartmann, Hollister, Lesniewski, Oakley, Parks and Robinson2009).

Fig. 7. Rarefaction curves based on bacterial 16S rRNA gene sequences from Hot Lake. The curves represent different levels of sequence identity.
Table 1. Diversity analyses of Hot Lake bacterial 16S rRNA gene sequences
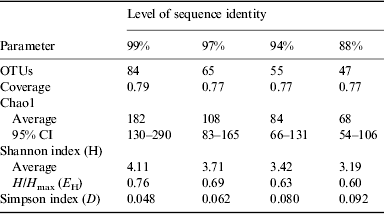
Rarefaction curves are presented in Fig. 7. None of these curves levels off, indicating that more sampling is needed to describe this community. Chao1 estimates were used to project the total number of OTUs at different levels of sequence identity (Table 1). At the 99% sequence identity level, 182 OTUs were predicted, while 84 OTUs were predicted at the 97% sequence identity level. It is estimated that 46, 60 and 65% of the bacterial diversity was sampled at 99, 97 and 94% sequence identity, respectively. The Shannon and Simpson indices both suggest that bacterial diversity increases at higher levels of sequence identity. The distribution of bacterial taxa in the library is relatively even since a measure of evenness, H/H max, is relatively high across all levels of identity. No equivalent epsomite environment has been described previously for direct comparison. The diversity estimates of the bacterial community at Hot Lake are similar to those of previous studies of environments rich in NaCl (Caton et al. 2004; Wani et al. Reference Wani, Surakasi, Siddharth, Raghavan, Patole, Ranade and Shouche2006; Mesbah et al. Reference Mesbah, Abou-El-Ela and Wiegel2007; Valenzuela-Encinas et al. Reference Valenzuela-Encinas, Neria-González, Alcántara-Hernández, Estrada-Alvarado, Zavala-Díaz de la Serna, Dendooven and Marsch2009).
Phylogenetic groups recovered
Phylogenetic trees based on the 16S rRNA clone library were generated that included sequences from cultured and uncultured bacteria obtained from the GenBank database (Figs 2, 3, 6 and S1–S3). Initial BLAST analyses were used to direct searches for rRNA gene sequences closely related to clones derived from Hot Lake. Clones from Gram-positive bacteria represented 40% of the library. Of these, clones related to uncultured actinomycetes dominated (>85%). Bacillus, Clostridia, Lactococcus and Staphylococcus also were detected. Staphylococcus is one of the few genera both cultivated and detected in the clone library.
Clones related to Gram-negative bacteria represented approximately 30% of the library. Legionella and Coxiella represented nearly half of the Gram-negative clones. Deltaproteobacteria were abundant in the clone library. This division is rich in sulphate-reducing bacteria that might be expected in an environment saturated in sulphate salts. Halothiobacillus, a purple sulphur bacterium in the Gammaproteobacteria, was observed and is presumably involved in autotrophic metabolism at Hot Lake. Clones related to Acidovorax, Erythrobacter and Lysobacter also were detected. Although Halomonas dominated the isolate collection, these were not detected in the clone library.
A number of clones clustered only with rRNA sequences from uncultured bacteria, some of which belong to candidate divisions. Approximately half of these clones clustered with members of candidate division TM6. A group of seven clones including HL2_C08 were not clearly related to known taxa. Clones related to TM7, OP10 and OP11 also were detected. A group of clones were related to the photosynthetic Chloroflexi.
Comparison of clone libraries from Hot Lake and JPL SAFs
A composite library of sequences obtained from SAF samples at JPL was compared to 277 sequences in the Hot Lake collection using BLAST. Approximately one third of the Hot Lake sequences matched the JPL SAF library at the genus (≥94% identity) level. At the species level (≥97% identity), approximately 13% of the Hot Lake sequences matched the JPL SAF library. The best matches were predominantly with Hot Lake isolates, especially Bacillus, Halomonas, Kocuria and Nocardiopsis. All Hot Lake sequences with less than 88% identity (division level) with the JPL SAF library were from clone libraries, not isolates. Both collections appear to be rich in microbes commonly found in soils including actinomycetes. An initial comparison with a composite sequence library from ESA SAFs showed less overlap with the Hot Lake collection as the ESA collection appeared to be richer in microbes typically associated with humans.
Discussion
Our recent paper reported that broad epsotolerance to 2 M MgSO4 is common in halotolerant bacteria from the GSP (Crisler et al. Reference Crisler, Newville, Chen, Clark and Schneegurt2012). Now we show that a natural environment with saturating concentrations of MgSO4 supports a diverse microbial community with broad epsotolerance and halotolerance. Greater halotolerance was generally observed in the GSP isolates than in Hot Lake isolates. Reciprocally, greater epsotolerance was generally observed in Hot Lake isolates than in GSP isolates. Within highly salinotolerant organisms apparently there is ionic specialization that concurs with their natural habitats to some extent. Nonetheless, substantial halotolerance was observed in isolates from Hot Lake, which does contain high concentrations of NaCl. It should be noted that while the Hot Lake bacterial isolates have broad halotolerance and epsotolerance, none were halophiles or epsophiles that require high salts for growth. At the GSP, the predominance of halotolerant organisms over halophilic organisms was suggested to have resulted from the rapidly changing salinity that accompanies flooding rain events and subsequent drying. At Hot Lake, broad salinotolerance may be a reflection of seasonal dilution with snowmelt and evaporative concentration in the warmer months.
Since no epsomite lake had been examined microbiologically, it was unknown whether bacterial isolates would be identified as those typically associated with hyperhaline environments, salinotolerant relatives of nearby forest soil bacteria, or bacteria endemic to epsomite environments. Most of the cultivated isolates and many of the molecular clones from Hot Lake were assigned to taxa commonly observed in hyperhaline environments such as Bacillus, Halomonas and Marinococcus. In contrast, Hot Lake yielded a number of actinomycetes that are not as widely reported from hyperhaline environments. No actinomycetes were isolated from the GSP, for example, using similar techniques. Actinomycetes are a numerically important cluster of bacterial clones from the culture-independent library. Initial deep sequencing of Hot Lake samples confirms the abundance of actinomycetes, in addition to a high number of Clostridia (Vaishampayan and Chen, unpublished). In fact, anaerobic bacteria appear to be common at Hot Lake with abundant sulphate-reducing bacteria in the clone library. Isolates and clones from the CFB group were abundant at the GSP, but absent in the Hot Lake isolate and clone collections. Some CFB group clones were detected in initial deep sequencing of Hot Lake. A significant portion of the clone library was assigned to unaffiliated taxa for which isolates have not been reported. While the isolate collection was dominated by bacteria typically associated with hyperhaline locations, the clone library more resembled a common soil community with abundant actinomycetes, Bacillus, Clostridia, Legionella and Acidovorax.
Perhaps the most surprising feature of the microbial community at Hot Lake was the near absence of archaea. In hyperhaline environments, archaea often dominate near saturating salinities. Archaeal 16S rRNA gene sequences were detected in certain Hot Lake samples by PCR, but these were in low abundance (Kilmer et al. Reference Kilmer, Eberl, Crisler, Cunderla, Madu and Schneegurt2012). One explanation is that many haloarchaea are extreme halophiles that require at least 1.5 M NaCl for growth (Mohr & Larsen Reference Mohr and Larsen1963; Schneegurt Reference Schneegurt and Vreeland2012). This salt requirement apparently cannot be fulfilled by other salts (Mullakhanbhai & Larsen Reference Mullakhanbhai and Larsen1975; Onishi et al. Reference Onishi, Fuchi, Konomi, Hidaka and Kamekura1980; Vreeland & Martin Reference Vreeland and Martin1980). Hot Lake does not have high concentrations of NaCl; so extremely halophilic archaea would not be expected to grow there. The archaea detected in sediments may be salinotolerant methanogens rather than haloarchaea.
It is becoming clear that epsotolerance is more widespread than one might expect given the limited distribution of epsomite environments on Earth. Initial studies of garden soils found a measurable number of epsotolerant and halotolerant microbes (Porazka et al. Reference Porazka, Kilmer and Schneegurt2011). This includes soil samples taken near the SAFs at JPL, increasing the potential for contamination of spacecraft with microbes tolerant to high MgSO4. Furthermore, initial analyses indicate that epsotolerant bacteria can be isolated from SAFs and clones from related taxa were detected using molecular means (Kilmer et al. Reference Kilmer, Eberl, Crisler, Cunderla, Madu and Schneegurt2012). Epsotolerance may increase the likelihood that a microbial contaminant could survive after a heat-producing crash landing of a spacecraft. Special habitats on Mars where liquid water is present may be heavy brines rich in sulphates of magnesium, calcium and iron, perhaps as eutectic liquids produced through deliquescence. Brines of perchlorate salts may be present in the north polar regions (McKay et al. Reference McKay, Stoker, Glass, Davé, Davila, Heldmann, Marinova, Fairen, Quinn and Zacny2013). Initial screening of bacterial isolates from Hot Lake and the GSP has shown considerable tolerance to perchlorates, in some cases, growing at 15% sodium or magnesium perchlorate (Mai et al. Reference Mai, Nosova and Schneegurt2012). A better understanding of terrestrial microbes growing under these conditions will impact life detection and sample return missions to Mars and other celestial bodies.
Acknowledgements
The authors are grateful for the preliminary and supportive work performed by Ashkaun Adib, James Crisler, John Dille, Felicia Giok, Jessica Pham, Kyle Rowe, Hutan Vahdat, Emily Winn and Lana Zayed. We are grateful to Stephen Lindemann (Pacific Northwest National Laboratory) for collecting Hot Lake samples and sharing related data. We thank Fadi Aramouni (Kansas State University) for performing water activity measurements, Christine Moissl-Eichinger (University of Regensburg) for sharing the ESA SAF bacterial 16S rRNA gene sequence database, and Parag Vaishampayan (NASA JPL) for comparing genetic libraries. This work was supported by awards from NASA ROSES Planetary Protection (PPR) and Kansas NASA EPSCoR. Additional support was provided by awards from NIH NCRR NIGMS KINBRE.
Supplementary materials
For supplementary material for this article, please visit http://dx.doi.org/10.1017/S1473550413000268.












