Abstract
It is widely agreed that the standard genetic code must have been preceded by a simpler code that encoded fewer amino acids. How this simpler code could have expanded into the standard genetic code is not well understood because most changes to the code are costly. Taking inspiration from the recently synthesized six-letter code, we propose a novel hypothesis: the initial genetic code consisted of only two letters, G and C, and then expanded the number of available codons via the introduction of an additional pair of letters, A and U. Various lines of evidence, including the relative prebiotic abundance of the earliest assigned amino acids, the balance of their hydrophobicity, and the higher GC content in genome coding regions, indicate that the original two nucleotides were indeed G and C. This process of code expansion probably started with the third base, continued with the second base, and ended up as the standard genetic code when the second pair of letters was introduced into the first base. The proposed process is consistent with the available empirical evidence, and it uniquely avoids the problem of costly code changes by positing instead that the code expanded its capacity via the creation of new codons with extra letters.
Similar content being viewed by others
Introduction
The standard genetic code is at the core of the procedure by which heritable differences are translated from genetic material, namely RNA, into amino acid sequences conforming proteins in living cells. It is known to be universal in basically all life forms, with minor exceptions (Bezerra et al. 2015; Knight et al. 2001). Protein synthesis consists in the successive translation of nucleotide triplets (codons) into one of twenty amino acids or into a stop signal.Footnote 1 During synthesis each codon will bond with the anticodon of its corresponding tRNA, which carries the amino acid assigned to the codon, via base pairing. For this reason the nucleotide letters of the genetic code come in pairs. Two such pairs exist, abbreviated by the letters guanine-cytosine (G-C) and adenine-uracil (A-U). In DNA, U is substituted by thymine (T).
The standard genetic code is robust against errors in translation and replication because of its highly regular arrangement of assignments from codons to amino acids (Freeland et al. 2003; Woese 1965). Even after over half a century of intensive investigations, and despite the development of several theories that attempt to explain the code’s origin in a rational manner, explaining how this code originated continues to be one of the major outstanding challenges of science (Koonin and Novozhilov 2009; Koonin and Novozhilov 2017). There is consensus that the code is too optimized to have emerged all at once, and that it was therefore likely preceded by a much simpler code that then expanded into the standard genetic code by incorporating more amino acids (Ikehara and Nihara 2007; Wong 2005).
However, most changes to the code are costly: an increase in the number of codon bases, such as from a doublet to the standard triplet (see, e.g., Copley et al. 2005), would turn all existing gene sequences into unreadable nonsense (Higgs 2009). More specifically, letters belonging to the first position of the next codon would be consistently misread as being the last letter of the preceding codon. In addition, even when assuming that the code began on the basis of triplet codons with lots of redundancy, subsequent reassignments of codons to new amino acids tend to disrupt protein function, especially when the newly assigned amino acids are chemically distant from previously assigned ones. Different solutions to this problem of costly code changes have been proposed, for example by balancing the deleterious consequences of codon reassignments with adaptive advantages of increased amino acid diversity (Higgs 2009).
However, there is another possibility of code evolution that manages to avoid this problem of costly changes altogether and that has not yet been sufficiently considered: an expansion of the number of codons available for amino acid assignment via an increase in the number of nucleotide letters employed by the three bases of a codon from two to four. The possibility of a primordial two-letter code had been proposed previously by Jiménez-Sánchez (1995) as a theoretical possibility, but recent advances in synthetic biology have demonstrated that increasing the number of letters employed by the code is in fact a practical possibility.
Synthetic biology has long been working on the creation of new life forms by artificially expanding the genetic code by re-allocating some codons to encode amino acids that are not in the naturally existing twenty (Xie and Schultz 2005). In an exciting new development, Zhang et al. (2017) were able to modify the standard genetic code of an organism such that it includes a third, unnatural base pair and its in vivo transcription into mRNA’s and into tRNA’s anticodons, resulting in an organism that incorporates natural and new amino acids, and thus encodes and retrieves increased heritable information. These results successfully turned the organism’s genetic code into a six- rather than a four-letter code.
Among other things, their work serves as a proof of concept that the number of letters of the primordial genetic code was not necessarily fixed right from its origin, but that it might have changed during its evolution. In particular, their work confirms that the introduction of additional bases into the genetic code has the effect of increasing its informational capacity, and that this can happen without disrupting the already established assignments of codons to amino acids and without producing nonsense out of existing gene sequences.
Importantly, this cost-free manner of increasing informational capability is a unique property of increasing the number of base pairs. In contrast, an increase in the number of codon bases from three to four, for example, would have also increased the informational capacity of the genetic code, by similarly increasing the number of available codons, but at the unsustainable cost of making all existing genome sequences that were coded in terms of triplets, unreadable and hence useless. The key implication of this contrast is that if the genetic code has increased its informational capacity during evolution, which is a reasonable assumption, then this would have most likely happened in terms of an increase in its nucleotide alphabet from two to four letters, rather than an increase in the number of codon bases. In line with Jiménez-Sánchez’ (1995) original proposal, this leads us to suggest that the standard genetic code could have started as a two-letter code before evolving into a four-letter code.
However, in most other respects our current proposal differs significantly from Jiménez-Sánchez’ original scenario. He argued that the primordial two-letter code was most likely based on the letters A and U, and that its eight codons were encoding one stop codon and the following seven amino acids: lysine (lys), asparagine (asn), tyrosine (tyr), methionine (met), isoleucine (ile), leucine (leu), and phenylalanine (phe). This is an odd scenario for various reasons.
First, this genetic code would have completely lacked redundancy at a stage in the evolution of life when redundancy was presumably needed most, namely in order to keep in check the deleterious effects of elevated rates of translation errors and mutations due to a still primitive genetic system. Second, the amino acid assignments are highly unlikely because many of them would have been rare or even absent in the environment at the origin of life. According to the analysis of likely prebiotic abundance of the encoded amino acids by Higgs and Pudritz (2009), three of them would have been absent altogether (asn, tyr, met), two would have been rare (lys, phe), and two would have been uncommon (leu, ile). Third, it is not clear whether an explicit stop signal would have even been needed at this early stage. Some theories have proposed that a simplified genetic code could have emerged before translation, and thus before stop codons would have been relevant (Copley et al. 2005). Alternatively, translation could have operated without stop codons as long as genes could have been encoded in short RNA fragments that broke off translation when their end was reached. Fourth, there are several reasons to doubt that a primordial two-letter code would have consisted of A and U. We now address this point in some detail.
To begin with, Jiménez-Sánchez argues that evolution would have favored the eventual introduction of the GC base pair because of its higher physicochemical stability compared to AU. Indeed, the GC bond is stronger because it is based on three hydrogen bonds, whereas the AU bond is based only on two. In addition, stability is also related to interactions of base stacking, that again favor GC (Yakovchuk et al. 2006). However, it seems more plausible that this extra stability provided by GC would have been most needed right at the origin of the genetic code rather than at a later stage, because initially the genetic system had not yet been fully optimized by evolution and thus plausibly relied more heavily on the stability of the base pairs to keep errors in check.
In particular, the GC bond is much more stable against the influence of high temperatures, and given that two of the most developed scenarios for the origin of life on earth envision this momentous event to have taken place either in deep-sea hydrothermal vents or in terrestrial hydrothermal fields (Deamer and Georgiou 2015; Djokic et al. 2017; Smith and Morowitz 2016), they both imply that GC was better suited than AU, at least until life moved to cooler habitats. Thus, it makes more sense to assume that the AU base pair was introduced at a later stage when the genetic system was sufficiently optimized to be able to compensate for its reduced stability.
Moreover, while Jiménez-Sánchez correctly observes that the frequency of an amino acid in modern proteins is positively correlated with the number of codons assigned to it, and also with its average GC/AU ratio, he takes this to support the precedence of AU over GC. His reasoning is that the higher codon redundancy that is characteristic of amino acids with higher GC content would make them less susceptible to being substituted in evolution. However, a recent simulation model of genetic code evolution has found that codon redundancy is also positively correlated with the frequency with which codons assigned to these amino acids are transferred between individual protocells (Froese et al. 2018a), which would instead turn increased codon redundancy into an indicator of evolutionary antiquity. Be that as it may, if the informational capacity of the genetic code expanded via an increase from two to four letters then there is no longer any need to assume that the evolution of the code involved costly codon reassignments in the first place.
Instead, the fact that the codons assigned to the most frequent amino acids in proteins have a higher GC content could indicate that the code had originally started as a two-letter GC code, and that it was only later complemented by the rarer amino acids, which consequently had less codons assigned and were more likely coded with the new letters AU. This is consistent with the fact that the most frequent amino acids in modern proteins also tend to be the ones most frequently found under prebiotic conditions. Finally, the possibility that the first two letters were G and C is additionally supported by the higher GC content of those regions of the genome with higher gene density (Pozzoli et al. 2008; Wuitschick and Karrer 1999).
In sum, while the general proposal that the genetic code evolved from a two- to a four-letter code has merit, it is evident that the previous proposal of an AU-code is problematic. In the following we sketch an alternative scenario of the initial form and subsequent evolution of this primordial two-letter genetic code, as well as remark upon the testability of this hypothesis.
One basic assumption of our hypothesis is that the later addition of A and U to the different bases of a codon did not change the assignments already made in the primordial code. Avoiding reassignments completely eradicates the costs of code evolution, and it also allows us to verify the consistency of our approach in terms of likely prebiotic abundances of the assigned amino acids. Secondly, we assume that the incorporation of the new pair of nucleotides occurred gradually, first by incorporating just one AU pair in each codon, then two and finally three. Finally, we can take into account Crick’s (1966) “wobble” effect, which means that the third position in the codon triplet plays a lesser defining role in the amino acid determination. It has been pointed out that this may be a vestigial effect of a two-base proto-code, which did not make meaningful use of the extant third base until later (Patel 2005; Travers 2006). There are independent reasons to assume that triplet codons were the most energetically efficient solution to the problem of reading gene sequences (Aldana-González et al. 2003), whether the third base made a difference to amino acid assignments or not.
We thus pose that at this initial stage the primordial code already had three-base codons, with a third-base wobble, but restricted to using a two-letter alphabet consisting only of G and C. We now assess the resulting implications.
Results
Initial GC-Only Code
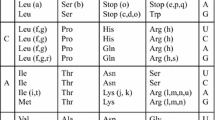
A primordial two-letter GC genetic code would have had the potential to specify a maximum of eight distinct elements (i.e. amino acids and stop codons). Based on the codon assignments of the standard genetic code and our basic assumptions, however, we find that this code consisted only of a subset of four amino acids (pro, ala, arg, and gly), as presented in Table 1.
Subsequent Code Expansion
The next phase of the evolution of the genetic code would have consisted in the gradual addition of the new AU base pair in different positions of the codon triplet. We assume that this code expansion would have occurred in a step-by-step manner. In other words, we first consider codons of the standard genetic code that differ from the original two-letter GC code in only one of the three positions. This subset of codon additions is shown in Table 2.
The next phase of genetic code evolution would have added codons that are yet further removed from the original two-letter GC code, namely by including A or U in two of the three possible positions of a codon (Table 3).
The last phase of genetic code evolution would have added codons that are even further removed from the original two-letter GC code, namely by including A or U in all three possible positions of a codon.
Consistency with Ranking of Prebiotic Amino Acid Abundance
One possibility of assessing the consistency of the primitive two-letter GC code and of the three subsequent phases of code expansion with existing evidence is to determine the average ranking of amino acid abundance. The proposed two-letter genetic code does not include amino acids that have not been associated with prebiotic contexts. We similarly expect that as the code evolves the average ranking of amino acid abundance will tend to decrease (recall that a ranking of 1 represents the most abundant, such that decreasing abundance correlates with a larger ranking number). In other words, later phases of code evolution are more likely to add amino acids that are increasingly less readily available in prebiotic contexts. And indeed the respective average rankings of amino acids encoded in the original GC code and those added during the three subsequent stages of code expansion (Tables 1, 2, 3 and 4) confirm this expectation, as illustrated in Fig. 1.
Average ranking of amino acid (AA) abundance in prebiotic contexts in terms of codon distance (0–3) from two-letter GC code. Y-axis: R_obs is the mean rank of amino acids observed in non-biological contexts (range [1.1–14.2]), as calculated by Higgs and Pudritz (2009); x-axis: sets of amino acids assigned to codons that are grouped according to their letter-based distance from the GC-code (0–3 letters). As expected, AAs that are assigned to codons that are more distant from the hypothesized primordial two-letter GC code tend to be less frequent or even altogether absent in prebiotic contexts. The whisker plots show the median value (orange line), the range of the 25% to the 75% quartile (blue box), and the range of the full dataset (black whiskers)
We can further refine our hypothesis by breaking down these four phases into smaller steps, given that not all three codon bases are equally important, as indicated by Crick’s wobble principle. It is possible to investigate which of the three positions of a codon is likely to have first expanded from two to four letters based on the likely prebiotic abundances of the amino acids. At this point it is insightful to reverse the procedure of the previous analysis, and to directly order all of the steps according to their average ranking. This results in a sequence of 8 steps, ordered as follows: 1) primordial GC code, 2) the third base of the codon accepts AU, 3) the second base accepts AU, 4) the second and third bases accept AU, 5) the first and third base accept AU, 6) the first base accepts AU, 7) the first, second, and third bases accept AU, and 8) The average rankings of prebiotic abundances associated with these steps are shown in Fig. 2.
Steps in the evolution from the two-letter to the four-letter code as predicted by the average ranking of amino acid abundance in prebiotic contexts (R_obs). Steps one and two and steps three and four have the same average ranking, while the average rankings of steps five to eight also form another cluster. The whisker plots show the median value (orange line), the range of the 25% to the 75% quartile (blue box), and the range of the full dataset (black whiskers). Outliers (circles) are outside their quartile limit by at least 1.5 times the interquartile range
A striking result of this ranking-based sequence of steps is the formation of three distinct clusters, corresponding with what would have been three distinct stages of changes to the code: Stage 1: the original GC code (code capacity: 8 codons) underwent a neutral change via the introduction of AU into the third base of the codons, although without any functional significance (code capacity: 16 codons). Stage 2: AU was introduced also into the second base (code capacity: 32 codons). Stage 3: AU was introduced also into the first base (code capacity: 64 codons). This is a plausible result and suggests that there were three major phases in the evolution of the genetic code, in which the originally all GC code was expanded by introducing AU letter-by-letter, moving from the third, to the second, to the first base, each time doubling the coding capacity of the code.
Consistency with Polar Requirement
Another test of this result of three stages of code expansion is to evaluate the distribution of polar requirements of the amino acids assigned to the available codons at each stage of code expansion. Hydrophobic amino acids have polar requirement values less than that of serine (7.5), while hydrophilic amino acids have larger values (Lenstra 2015). We expect that at each stage of code expansion the average polar requirement value will be close to this neutral value, which would indicate a balance between codons assigned to hydrophobic and to hydrophilic amino acids. This balance is important because binary patterning of amino acids with polar and nonpolar residues enhances the process of protein folding (Kamtekar et al. 1993).
The polar requirement values associated with each of the three stages of code expansion that we identified in the previous analysis is shown in Fig. 3. As we expected, the median value of polar requirement stays remarkably close to neutral (7.45, 7.45, and 7 for Stages 1–3, respectively), even as the overall range and diversity of values expands.
Polar requirements of amino acids encoded by all codons available at each stage of code expansion from two- to four-letter code. A polar requirement value of 7.5 is neutral, while hydrophobic amino acids have smaller values and hydrophilic amino acids have larger values. As expected, the amino acids that are assigned to the codons available at each predicted stage of code expansion are remarkably balanced in terms of their polar requirements, with median values of 7.45, 7.45, and 7 for Stages 1–3, respectively, even as the overall range of polar requirement values increases. Outliers (circles) are outside their quartile limit by at least 1.5 times the interquartile range
In summary, starting with a GC-code leads to a plausible scenario of subsequent code expansion. Moreover, we emphasize again that the key advantage of this scenario is that it can account for this expansion while sidestepping the usual negative consequences. First, by starting with codons that already include all three positions, there is no need to deal with the loss of genetic information that would have been caused by gene sequences becoming unreadable if the code had expanded by increasing the number of codon bases. Second, by starting with two letters rather than the full four, there is no need to deal with the constraints and malfunctions associated with codon reassignments (Higgs 2009). In other words, the form of code expansion we have proposed is much more evolvable than that of alternative scenarios found in the literature.
Discussion
A couple of features of the initial two-letter GC genetic code shown in Table 1 are worth noting. First, this GC-only codon subset of the standard genetic code does not make full use of the eight distinct amino acids or stop codons that could be assigned. Instead it includes only four amino acids, each with a redundancy of two codons. This arrangement maximizes the coding capacity while each amino acid has a minimum amount of redundancy. The GC-code therefore retains an ordered layout with “wobble-like” degeneracy (i.e. the third codon does not affect the amino acid assignments), which would have increased robustness against errors in copying and translation already at this early stage of code evolution. It is also consistent with existing proposals that the genetic code might have originally only made use of the first two positions while ignoring the third.
Second, according to the systematic review of the literature by Higgs and Pudritz (2009), all four amino acids encoded at this stage have been found in relevant non-biological contexts, including hydrothermal vents and meteorites, as well as in experiments designed to be informative about the conditions that likely held at the origins of life. In fact, when the twenty amino acids of the standard genetic code are ranked in order of decreasing abundance in these prebiotic contexts, glycine and alanine are consistently reported as number one and two most abundant, respectively. Proline, on the other hand, is found in most of these contexts, albeit in less concentration.
Somewhat surprisingly, this code would have also included the rather low-ranked arginine, a large and complex amino acid. Arginine has been regularly documented in specific experimental contexts involving synthesis from CO, H2, and NH3 at high temperature in the presence of catalysts, including clay (Yoshino et al. 1971).Footnote 2 Accordingly, if our proposal is on the right track, the implication is that those experiments that generated arginine deserve special attention, as they might hold clues with respect to the conditions under which the two-letter genetic code emerged. For instance, more attention should be paid to the kinds of amino acids that can be produced in hydrothermal pools and geysers (e.g. Damer and Deamer 2015; Djokic et al. 2017). It is also suggestive that arginine basically consists of glycine, which we assume was encoded by the GC code, plus a side chain that ends in a guanidine group, which can be derived from the degradation of guanine, i.e. precisely from the nucleotide G of the GC code.
A few features are also worth noting about the first phase of code expansion, which added those codons that differed from the primordial GC code in one position. First, and most importantly, this is an expansion of the two-letter GC code to the full four-letter basis of the standard genetic code, albeit in a spatially restricted way. Recall that the key advantage is that this inclusion of either A or U in one of the three positions increases the code’s information capability, but without turning existing gene sequences, that had originally evolved on the basis of the two-letter code, into nonsense. Thus, the incorporation of this batch of codons has had no effect on the assignments of the original GC-code. Second, all four amino acids that were specified in the original two-letter code have received further codon assignments, all of them when the new nucleotide occupies the third codon position. This result is again consistent with the “wobble” interpretation.
During this first expansion phase nine new amino acids were included in the expanded code, most of which have been associated with prebiotic conditions. However, there are three exceptions to this: glutamine, cysteine, and tryptophan. Together with histamine, which has a very low prebiotic presence, they constitute the only four “singlets” (non-degenerate codons) at this stage. This is remarkable and may lend support to the idea of a correlation between degeneracy and primordial abundance, which has been pointed out before in the context of non-uniform usage of synonymous codons (Satapathy et al. 2017). This also suggests a third consideration, namely that this and further steps in the expansion of the basis of the genetic code from two to four nucleotides may have occurred after the biological context had sufficient complexity to generate these amino acids, as for example proposed by the coevolution theory of the genetic code (Di Giulio 2008; Wong 2005).
The second phase of code expansion, which incorporated codons that differed from the GC code in two of the three bases of a codon, added seven new amino acids to the early genetic code (Ile, Lys, Phe, His, Asn, Met, Tyr), the last three of which have not been found in association with any prebiotic contexts or origin of life experiments. Again, these are non-redundant codons up to this evolutionary stage. We remark that this is also the phase at which stop codons make their first appearance, together with the methionine codon, which also plays the role of initiation or start codon, thus setting the stage for complex protein assembly.
In the final phase of code expansion, which incorporated codons that differed from the GC code in all three bases, there are no new amino acid additions to the genetic code. Nevertheless, a higher degeneracy is achieved and a new stop sign is included. After this phase the full 64 codons of the standard genetic code are recovered. The fact that codons consisting of only AU do not add any new amino acids is interesting, and may suggest that this phase actually co-occurred with the introduction of AU in codon positions 1 and 2. Indeed, when we take into account the specific codon base positions in which the expansion from a two-letter to four-letter occurs, and then use the estimates of the prebiotic abundance of the assigned amino acids as a way of ordering the steps of code evolution, we end up with three clusters of steps (Fig. 2). This suggests an even simpler scenario of code expansion consisting of only three rather than four phases:
Phase One (Gly, Ala, Pro, Arg)
This phase includes the primordial two-letter GC code (see Table 1), as well as those codons of the standard genetic code that differ from the two-letter code only in the third position. This grouping is consistent with the fact that the third position is less reliable (Crick’s “third base wobble”), and therefore might not have initially played any coding role. The third position was thus a suitable starting point for introducing new nucleotides.
Phase Two (Gly, Ala, Pro, Arg, Asp, Glu, Val, Leu, His, Gln)
This phase adds codons of the standard genetic code that have A or U only in the second position, and those that have A or U both in the second and in the third position. This grouping is consistent with Higgs and Pudritz’s (2009) observation that the five earliest amino acids according to their ranking (Gly, Ala, Asp, Glu, Val) all have a first position base G, which they interpret to suggest that the second base was the most important discriminator in the early code. In other words, during this stage the informational content of the code was increased via the addition of a new pair of letters to the second position. The third position continued to accept A and U in combination with the expansion of the second position but, as in stage one, the third position still played no coding role.
Phase Three (Gly, Ala, Pro, Arg, Asp, Glu, Val, Leu, His, Gln, Ser, Thr, Cys, Trp, Ile, Lys, Phe, Tyr, Asn, Met)
In this final stage of code evolution the first position of a codon also started to include A and U, and this offered new combinations with the other positions. The final four steps in Fig. 2 are ordered as follows: 1) AU in the first and third positions; 2) AU in the first position; 3) AU in the first, second, and third positions; and 4) AU in the first and second positions. This sequence of steps, however, cannot really be discriminated by prebiotic abundances. At this point it is better to treat them as one coherent group of changes. Nevertheless, it is worth noting that the codons of the three stop codons are distributed across steps 1), 3) and 4).
Our proposal receives further support from the fact that the median polar requirement value is maintained near the neutral value of 7.5 even as the overall diversity of values increases. Future work could look at these stages in terms of other relevant chemical properties. However, given that there does not seem to be a significantly different coverage of chemical property space when comparing high-ranked versus low-ranked amino acids (Froese et al. 2018b), we expect that balance is maintained more generally.
Finally, we note that a potential problem for our proposal arises if it is assumed that the primordial GC-code likely arose after large RNAs, like the ribosome, tRNAs, and mRNAs, were already present. This is because these RNAs consist of all four letters of the standard code and consequently it would be strange if life had a way of replicating these macromolecules but based its genetic code on only two of the available four letters.Footnote 3 To some extent this issue may put our proposal at odds with the “RNA world” hypothesis about the origin of life, which is viewed as the best game in town by many researchers in the field (Higgs and Lehman 2015). Nevertheless, this hypothesis still faces major unsolved issues, including precisely the problem of explaining the origin of the genetic code (Pressman et al. 2015). Moreover, it is not clear whether it is even necessary to assume that these large RNAs must have been present before the origin of the code. For example, Copley et al. (2005) have proposed that an association between 14 amino acids and their codons (or at least their first two bases) arose before the emergence of the macromolecules used by the RNA translation apparatus (see also Rodin et al. 2011). Relatedly, Froese et al. (2018a, 2018b) have shown with a simple simulation model of small groups of interacting protocells that the characteristic regularities of assignments between amino acids and codons of the standard genetic code can self-organize even in the absence of optimization via evolution by vertical descent. Starting with a GC code would have also aided the transition to a more complex arrangement involving four-letter RNA and DNA, because it avoids the need to deal with the difference between U (RNA) and T (DNA). We therefore think that it is an interesting open question whether the emergence of a GC-code could have preceded life’s capacity for replication of large RNAs that consisted of all four letters.
Conclusions
Our novel hypothesis that the standard genetic code had a binary, two-letter predecessor consisting of only G and C has merit and deserves further attention. The hypothesis has the crucial advantage that it can account for the main phases of genetic code expansion without creating nonsense of existing gene sequences and without having to deal with the problem of costly codon reassignments. This hypothesis therefore uniquely resolves a key challenge of current research into the origins and evolution of the genetic code. Moreover, the evolutionary route from this two-letter GC code to the standard four-letter code fits well with the available evidence: the primordial GC code is especially stable against the high temperatures expected by terrestrial origins of life scenarios; the sequence of expansion phases correlates with a decrease in relative prebiotic abundances of the newly encoded amino acids; the sequence exhibits redundancy and balanced polar requirements throughout all phases, and it includes stop codons only in the final phases of code expansion.
Finally, this hypothesis points to new avenues for research. Future work could, for example, determine whether some of the proposed early additions of amino acids that are somewhat less expected given the best current evidence about their prebiotic abundance could have been more common in contexts that have received less attention, such as hydrothermal pool scenarios. The hypothesis also leads to the testable prediction that the evolutionary age of contemporary gene sequences positively correlates with their GC content.
Notes
There are exceptions to this rule; certain microorganisms translate codons into the standard twenty amino acids plus an extra amino acid (Atkins and Gesteland 2002).
Proline can also emerge under these conditions, albeit not as regularly.
We thank Paul Higgs for bringing this issue to our attention.
Abbreviations
- A:
-
Adenine
- AA:
-
Amino acid
- Ala:
-
Alanine
- Arg:
-
Arginine
- Asn:
-
Asparagine
- Asp:
-
Aspartate
- C:
-
Cytosine
- Cys:
-
Cysteine
- DNA:
-
Deoxyribonucleic acid
- G:
-
Guanine
- Glu:
-
Glutamate
- Gln:
-
Glutamine
- Gly:
-
Glycine
- His:
-
Histidine
- Ile:
-
Isoleucine
- Leu:
-
Leucine
- Lys:
-
Lysine
- Met:
-
Methionine
- mRNA:
-
Messenger RNA
- RNA:
-
Ribonucleic acid
- R_obs:
-
Mean rank of concentrations of AAs observed in non-biological contexts
- Phe:
-
Phenylalanine
- Pro:
-
Proline
- Ser:
-
Serine
- T:
-
Thymine
- Thr:
-
Threonine
- tRNA:
-
Transfer RNA
- Trp:
-
Tryptophan
- Tyr:
-
Tyrosine
- U:
-
Uracil
- Val:
-
Valine
References
Aldana-González M, Cocho G, Larralde H, Martínez-Mekler G (2003) Translocation properties of primitive molecular machines and their relevance to the structure of the genetic code. J Theor Biol 220:27–45
Atkins JF, Gesteland R (2002) The 22nd amino acid. Science 296:1409–1410
Bezerra AR, Guimarães AR, Santos MAS (2015) Non-standard genetic codes define new concepts for protein engineering. Life 5:1610–1628. https://doi.org/10.3390/life5041610
Copley SD, Smith E, Morowitz HJ (2005) A mechanism for the association of amino acids with their codons and the origin of the genetic code. Proc Natl Acad Sci U S A 102:4442–4447
Crick FHC (1966) Codon-anticodon pairing: The wobble hypothesis. J Mol Biol 19:548–555
Damer B, Deamer D (2015) Coupled phases and combinatorial selection in fluctuating hydrothermal pools: A scenario to guide experimental approaches to the origin of cellular life. Life 5:872–887
Deamer DW, Georgiou CD (2015) Hydrothermal conditions and the origin of cellular life. Astrobiol 15:1091–1095
Di Giulio M (2008) An extension of the coevolution theory of the origin of the genetic code. Biol Direct 3. https://doi.org/10.1186/1745-6150-3-37
Djokic T, Van Kranendonk MJ, Campbell KA, Walter MR, Ward CR (2017) Earliest signs of life on land preserved in ca. 3.5 Ga hot spring deposits. Nat Commun 8:15263. https://doi.org/10.1038/ncomms15263
Freeland SJ, Wu T, Keulmann N (2003) The case for an error minimizing standard genetic code. Orig Life Evol Biosph 33:457–477
Froese T, Campos JI, Fujishima K, Kiga D, Virgo N (2018a) Horizontal transfer of code fragments between protocells can explain the origins of the genetic code without vertical descent. Sci Rep 8:3532. https://doi.org/10.1038/s41598-018-21973-y
Froese T, Campos JI, Virgo N (2018b) An iterated learning approach to the origins of the standard genetic code can help to explain its sequence of amino acid assignments. In: Ikegami T, Virgo N, Witkowski O, Oka M, Suzuki R, Iizuka H (eds) Proceedings of the Artificial Life Conference 2018. MIT Press, Cambridge
Higgs PG (2009) A four-column theory for the origin of the genetic code: Tracing the evolutionary pathways that gave rise to an optimized code. Biol Direct 4. https://doi.org/10.1186/1745-6150-4-16
Higgs PG, Lehman N (2015) The RNA World: Molecular cooperation at the origins of life. Nat Rev Genet 16:7–17
Higgs PG, Pudritz RE (2009) A thermodynamic basis for prebiotic amino acid synthesis and the nature of the first genetic code. Astrobiol 9:483–490
Ikehara K, Nihara Y (2007) Origin and evolutionary process of the genetic code. Curr Med Chem 14:3221–3231
Jiménez-Sánchez A (1995) On the origin and evolution of the genetic code. J Mol Evol 41:712–716
Kamtekar S, Schiffer JM, Xiong H, Babik JM, Hecht MH (1993) Protein design by binary patterning of polar and nonpolar amino acids. Science 262:1680–1685
Knight RD, Freeland SJ, Landweber LF (2001) Rewiring the keyboard: Evolvability of the genetic code. Nat Rev Genet 2:49–58
Koonin EV, Novozhilov AS (2009) Origin and evolution of the genetic code: The universal enigma. IUBMB Life 61:99–111
Koonin EV, Novozhilov AS (2017) Origin and evolution of the universal genetic code. Annu Rev Genet 51:45–62
Lenstra R (2015) The graph, geometry and symmetries of the genetic code with Hamming metric. Symmetry 7:1211–1260
Patel A (2005) The triplet genetic code had a doublet predecessor. J Theor Biol 233:527–532
Pozzoli U et al (2008) Both selective and neutral processes drive GC content evolution in the human genome. BMC Evol Biol 8:99. https://doi.org/10.1186/1471-2148-8-99
Pressman A, Blanco C, Chen IA (2015) The RNA World as a model system to study the origin of life. Curr Biol 25:R953–R963
Rodin AS, Szathmáry E, Rodin SN (2011) On origin of genetic code and tRNA before translation. Biol Direct 6. https://doi.org/10.1186/1745-6150-6-14
Satapathy SS, Sahoo AK, Ray SK, Ghosh TC (2017) Codon degeneracy and amino acid abundance influence the measures of codon usage bias: improved Nc (N̂c) and ENCprime (N̂′c) measures. Genes Cells 22:277–283
Smith E, Morowitz H (2016) The Origin and Nature of Life on Earth: The Emergence of the Fourth Geosphere. Cambridge University Press, Cambridge
Travers A (2006) The evolution of the genetic code revisited. Orig Life Evol Biosph 36:549–555
Woese CR (1965) On the evolution of the genetic code. Proc Natl Acad Sci U S A 54:1546–1552
Wong JT-F (2005) Coevolution theory of the genetic code at age thirty. BioEssays 27:416–425
Wuitschick JD, Karrer KM (1999) Analysis of genomic G + C content, codon usage, initiator codon context and translation termination sites in Tetrahymena thermophila. J Eukaryot Microbiol 46:239–247
Xie J, Schultz PG (2005) Adding amino acids to the genetic repertoire. Curr Opin Chem Biol 9:548–554
Yakovchuk P, Protozanova E, Frank-Kamenetskii MD (2006) Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res 34:564–574
Yoshino D, Hayatsu R, Anders E (1971) Origin of organic matter in early solar system - III. Amino acids: catalytic synthesis. Geochim Cosmochim Acta 35:927–938
Zhang Y et al (2017) A semi-synthetic organism that stores and retrieves increased genetic information. Nature 551:644–647
Acknowledgements
T.F. was supported by UNAM-DGAPA-PAPIIT project IA104717. We thank Michel Naslavsky and Karo Michaelian for their helpful comments on an earlier draft of this paper, and Jorge I. Campos for his help with the figures. We also thank Paul Higgs for his insightful review.
Author information
Authors and Affiliations
Contributions
AF conceived of a two-letter predecessor of the four-letter standard genetic code, and discussed the design, results and implications of the analysis. TF wrote the paper and performed the analysis. Both authors read and approved the final manuscript.
Corresponding author
Rights and permissions
About this article
Cite this article
Frank, A., Froese, T. The Standard Genetic Code can Evolve from a Two-Letter GC Code Without Information Loss or Costly Reassignments. Orig Life Evol Biosph 48, 259–272 (2018). https://doi.org/10.1007/s11084-018-9559-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11084-018-9559-4